蛋白质工程中基于生物物理学的蛋白质语言模型。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
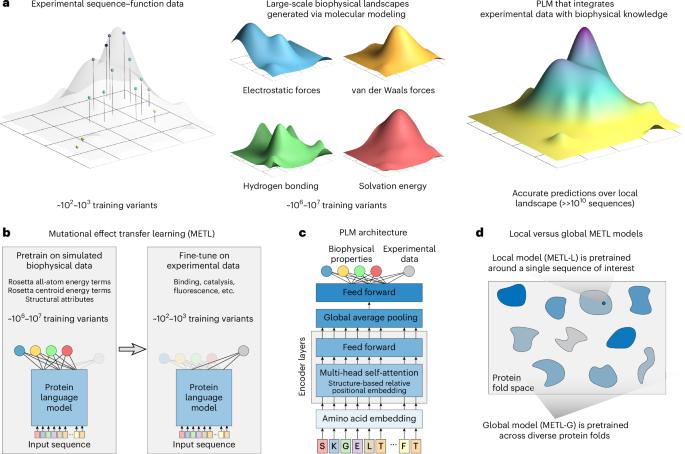
经过进化数据训练的蛋白质语言模型已经成为预测涉及蛋白质序列、结构和功能问题的强大工具。然而,这些模型忽略了几十年来对控制蛋白质功能的生物物理因素的研究。我们提出突变效应迁移学习(METL),这是一种结合先进机器学习和生物物理建模的蛋白质语言模型框架。利用METL框架,我们基于生物物理模拟数据预训练基于变压器的神经网络,以捕获蛋白质序列、结构和能量学之间的基本关系。我们对METL在实验序列功能数据上进行微调,以利用这些生物物理信号,并将其应用于预测蛋白质特性,如热稳定性、催化活性和荧光。METL擅长于挑战性的蛋白质工程任务,如从小训练集进行归纳和位置外推,尽管现有的训练进化信号的方法在许多类型的实验分析中仍然很强大。我们展示了METL在64个样本上训练后设计功能性绿色荧光蛋白变体的能力,展示了基于生物物理学的蛋白质语言模型在蛋白质工程中的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Biophysics-based protein language models for protein engineering
Protein language models trained on evolutionary data have emerged as powerful tools for predictive problems involving protein sequence, structure and function. However, these models overlook decades of research into biophysical factors governing protein function. We propose mutational effect transfer learning (METL), a protein language model framework that unites advanced machine learning and biophysical modeling. Using the METL framework, we pretrain transformer-based neural networks on biophysical simulation data to capture fundamental relationships between protein sequence, structure and energetics. We fine-tune METL on experimental sequence–function data to harness these biophysical signals and apply them when predicting protein properties like thermostability, catalytic activity and fluorescence. METL excels in challenging protein engineering tasks like generalizing from small training sets and position extrapolation, although existing methods that train on evolutionary signals remain powerful for many types of experimental assays. We demonstrate METL’s ability to design functional green fluorescent protein variants when trained on only 64 examples, showcasing the potential of biophysics-based protein language models for protein engineering. Mutational effect transfer learning (METL) is a protein language model framework that unites machine learning and biophysical modeling. Transformer-based neural networks are pretrained on biophysical simulation data to capture fundamental relationships between protein sequence, structure and energetics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


