拟南芥氨基转移酶的多底物特异性定位
IF 13.6
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
摘要
氮是所有生物必需的元素,其有效性和利用效率直接影响生物的生长和性能,特别是在植物中。转氨酶是合成各种有机氮化合物的氮代谢网络的核心酶。尽管每种转氨酶都可能催化数百种不同氨基和酮酸底物组合的转氨化反应,但许多转氨酶的完整功能仍然难以捉摸。本文采用高通量基因合成和酶分析平台确定了拟南芥38种转氨酶的底物特异性,并在总共4,104个反应中揭示了许多以前未被认识的活性。将这些生化数据整合到拟南芥酶约束代谢模型和硅模拟中,进一步揭示了转氨酶的乱交可能改变氮的分布特征,并有助于氮代谢网络的鲁棒性。该研究为破解植物氮素代谢网络,提高作物氮素利用效率提供了基础知识。本文章由计算机程序翻译,如有差异,请以英文原文为准。
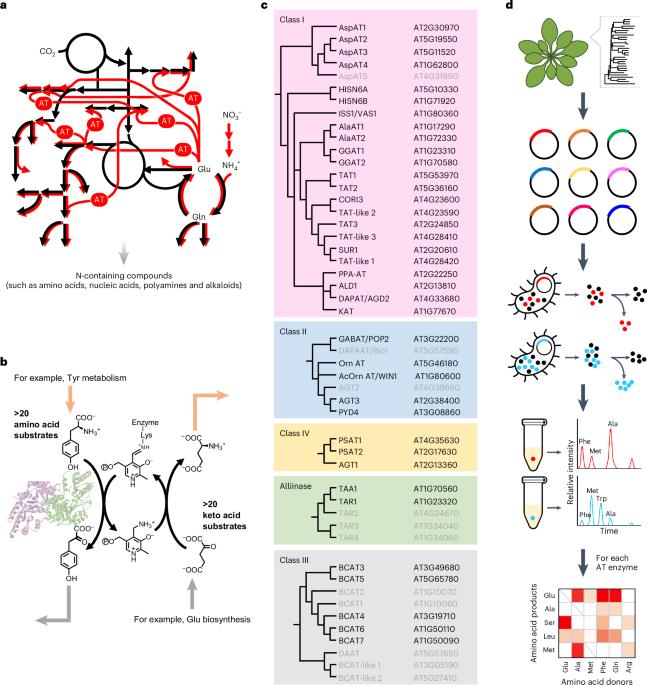
Mapping multi-substrate specificity of Arabidopsis aminotransferases
Nitrogen is an essential element in all organisms, and its availability and use efficiency directly impact organismal growth and performance, especially in plants. Aminotransferases are core enzymes of the nitrogen metabolic network for synthesizing various organonitrogen compounds. Although each aminotransferase can potentially catalyse hundreds of transamination reactions with different combinations of amino and keto acid substrates, the full functionality of many aminotransferases remains elusive. Here we employed high-throughput gene synthesis and enzyme assay platforms to determine the substrate specificities of 38 aminotransferases of Arabidopsis thaliana and unveiled many previously unrecognized activities among a total of 4,104 reactions tested. The integration of these biochemical data in an enzyme-constrained metabolic model of Arabidopsis and in silico simulation further revealed that the promiscuity of aminotransferases may alter nitrogen distribution profiles and contribute to the robustness of the nitrogen metabolic network. This study provides foundational knowledge for deciphering the plant nitrogen metabolic network and improving nitrogen use efficiency in crops. Systematic characterization of Arabidopsis aminotransferase family enzymes uncovered many previously unrecognized activities and revealed their multi-substrate specificity, aspects that probably contribute to the robustness of the nitrogen metabolic network.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


