口腔微生物组研究中核酸保存方案的评价。
IF 1.9
4区 生物学
Q4 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
目的:口腔微生物组研究的准确性在很大程度上取决于标本取样方案,以及它们的储存和保存。传统的方法,如冷冻,可能不仅涉及后勤障碍,而且可能影响微生物数据的质量,导致不同研究之间的可比性困难。本研究评估了使用DNA/RNA屏蔽缓冲液的室温核酸保存方案的有效性,与标准冷冻相比,在7 天的过程中保存口腔微生物群落。结果:基于16S rRNA基因测序的比较分析显示,两种保存策略获取的微生物组数据总体上具有高度一致性。在DNA产率方面,qPCR分析显示,使用DNA/RNA Shield时,从舌拭子、牙袋和唾液样本中回收的细菌DNA显著增加。在分类学分析中,与冷冻相比,Shield缓冲液保存了更高丰度的细孔菌。然而,对于所有其他分类群,两种方案之间的组成数据具有高度可比性。虽然两种保存方法对大多数样品的微生物多样性没有明显差异,但DNA/RNA Shield中保存的唾液样品的多样性明显更高。这两种保存方法在检索一致的生物学读数(如位点特异性和健康状态)的能力方面具有高度可比性。结论:室温下用DNA/RNA屏蔽缓冲液保存口腔样品是一种可靠、有效的微生物组保存方法。此外,它可以提高DNA产量,同时保持样品中的微生物多样性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
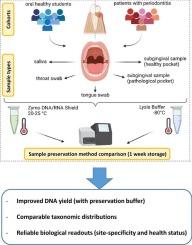
Evaluation of a nucleic acid preservation protocol for microbiome studies involving samples from the oral cavity
Purpose
The accuracy of oral microbiome research depends significantly on specimen sampling protocols, as well as their storage and preservation. Traditional methods, such as freezing, may not only involve logistical hurdles but can also impact the quality of microbial data, leading to difficulties in the comparability between different studies. This study evaluates the effectiveness of the room temperature nucleic acid preservation protocol using DNA/RNA Shield buffer as compared to standard freezing in preserving oral microbial communities over the course of 7 days.
Results
Comparative analyses based on 16S rRNA gene sequencing revealed a high overall consistency between the microbiome data retrieved by both preservation strategies. In terms of DNA yield, qPCR analysis showed a significant increase in bacterial DNA recovered from tongue swabs, dental pockets and saliva samples when using DNA/RNA Shield. In the taxonomic analyses, the Shield buffer preserved a higher abundance of Veillonella as compared to freezing. However, the compositional data was highly comparable between both protocols for all other taxonomic groups. While no major differences in microbial diversity were observed between both preservation methods for most samples, saliva samples stored in DNA/RNA Shield exhibited significantly higher diversity. Both preservation methods performed in a highly comparable manner in their ability to retrieve consistent biological readouts, such as site-specificity and health status.
Conclusion
Storing oral samples at room temperature in DNA/RNA Shield buffer provides a reliable and efficient alternative to freezing for preserving microbiome samples. Furthermore, it may enhance DNA yield while preserving microbial diversity in the samples.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of microbiological methods
生物-生化研究方法
CiteScore
4.30
自引率
4.50%
发文量
151
审稿时长
29 days
期刊介绍:
The Journal of Microbiological Methods publishes scholarly and original articles, notes and review articles. These articles must include novel and/or state-of-the-art methods, or significant improvements to existing methods. Novel and innovative applications of current methods that are validated and useful will also be published. JMM strives for scholarship, innovation and excellence. This demands scientific rigour, the best available methods and technologies, correctly replicated experiments/tests, the inclusion of proper controls, calibrations, and the correct statistical analysis. The presentation of the data must support the interpretation of the method/approach.
All aspects of microbiology are covered, except virology. These include agricultural microbiology, applied and environmental microbiology, bioassays, bioinformatics, biotechnology, biochemical microbiology, clinical microbiology, diagnostics, food monitoring and quality control microbiology, microbial genetics and genomics, geomicrobiology, microbiome methods regardless of habitat, high through-put sequencing methods and analysis, microbial pathogenesis and host responses, metabolomics, metagenomics, metaproteomics, microbial ecology and diversity, microbial physiology, microbial ultra-structure, microscopic and imaging methods, molecular microbiology, mycology, novel mathematical microbiology and modelling, parasitology, plant-microbe interactions, protein markers/profiles, proteomics, pyrosequencing, public health microbiology, radioisotopes applied to microbiology, robotics applied to microbiological methods,rumen microbiology, microbiological methods for space missions and extreme environments, sampling methods and samplers, soil and sediment microbiology, transcriptomics, veterinary microbiology, sero-diagnostics and typing/identification.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


