基因编码FerriTag作为冷冻电子断层扫描的特定标签
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
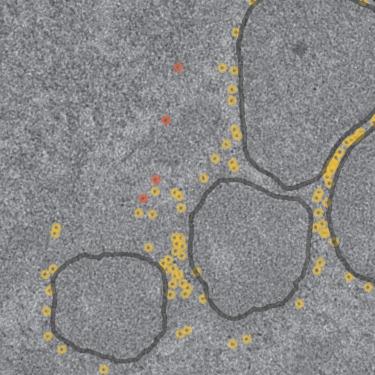
低温电子断层扫描(cryoET)提供纳米和亚纳米分辨率的细胞器和蛋白质的3D数据集。然而,在活细胞中定位靶蛋白仍然是一个重大挑战。传统的标记方法,如荧光蛋白标记和免疫金标记,不适合在分子分辨率下玻璃化样品中的小结构。直接连接大的,视觉上可识别的蛋白质到目标蛋白质可能会改变它们的结构,定位和功能。为了克服这个问题,我们采用了一种雷帕霉素诱导的低聚物形成系统,包括两个标签,FK506结合蛋白(FKBP)和FKBP-雷帕霉素结合(FRB),它们在雷帕霉素存在下结合。FKBP与靶蛋白相连,而FRB与铁蛋白相连,铁蛋白是一种大的(10-12纳米)铁结合复合物,在低温低温下形成强烈的对比。在细胞培养基中加入雷帕霉素后,载铁铁蛋白准确地标记出目标蛋白的位置。随着亚层析成像平均技术的进步,我们的方法解决了在活细胞中定位目标蛋白的持续挑战。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Genetically encoded FerriTag as a specific label for cryo-electron tomography
Cryo-electron tomography (cryoET) provides 3D datasets of organelles and proteins at nanometer and sub-nanometer resolution. However, locating target proteins in live cells remains a significant challenge. Conventional labeling methods, such as fluorescent protein tagging and immunogold labeling, are unsuitable for small structures in vitrified samples at molecular resolution. Directly linking large, visually identifiable proteins to target proteins may alter their structure, localization, and function. To overcome this, we employed a rapamycin-induced oligomer formation system involving two tags, FK506 binding protein (FKBP) and FKBP-rapamycin binding (FRB), which bind in the presence of rapamycin. FKBP is linked to the target protein, while FRB is linked to ferritin, a large (10–12 nm) iron-binding complex that creates strong contrast in cryoET. Upon adding rapamycin to the cell medium, the iron-loaded ferritin accurately marks the target protein location. As in situ cryoET with subtomogram averaging advances, our method addresses the persistent challenge of locating target proteins in live cells.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


