rna结合蛋白介导分化过程中染色质拓扑结构的成熟
IF 19.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
摘要
拓扑关联结构域(TAD)和染色质结构环影响启动子-增强子的相互作用,ccctc结合因子(CTCF)定义TAD边界和环锚点。TAD边界和环在胚胎干细胞分化过程中逐渐加强,强调了染色质拓扑结构在个体发生中的重要性。然而,推动这一过程的机制仍不清楚。在这里,我们发现ctcf - rna结合蛋白(RBP)相互作用在ES向神经干细胞(NS)分化过程中广泛增加。虽然在胚胎干细胞中是不可缺少的,但在NS细胞中,rbp增强了ctcf锚定的染色质拓扑结构。我们发现非编码RNA Pantr1是CTCF-RBP相互作用的关键促进物,促进染色质成熟。利用急性CTCF降解,我们发现,通过其绝缘体功能,CTCF作为发育过程中过早基因激活的屏障,有助于维持NS细胞中的神经元基因沉默。总之,我们揭示了一种驱动发育相关染色质结构巩固的基本机制,以及这一过程对分化中基因表达控制的贡献。本文章由计算机程序翻译,如有差异,请以英文原文为准。
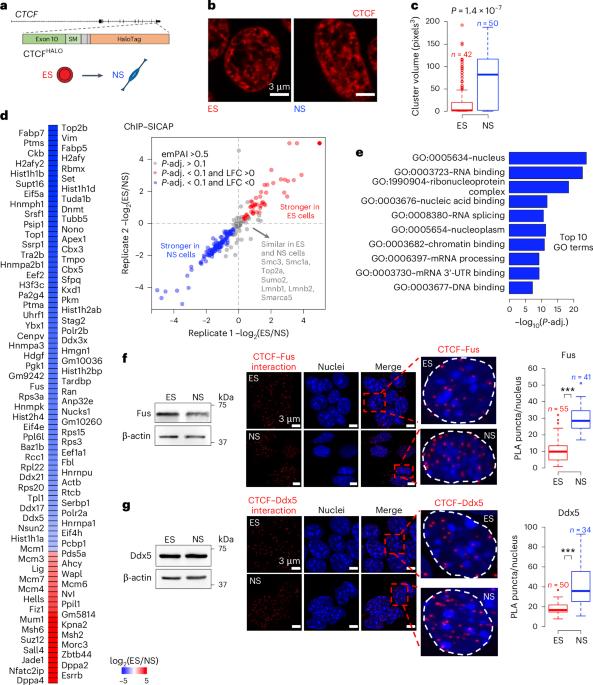
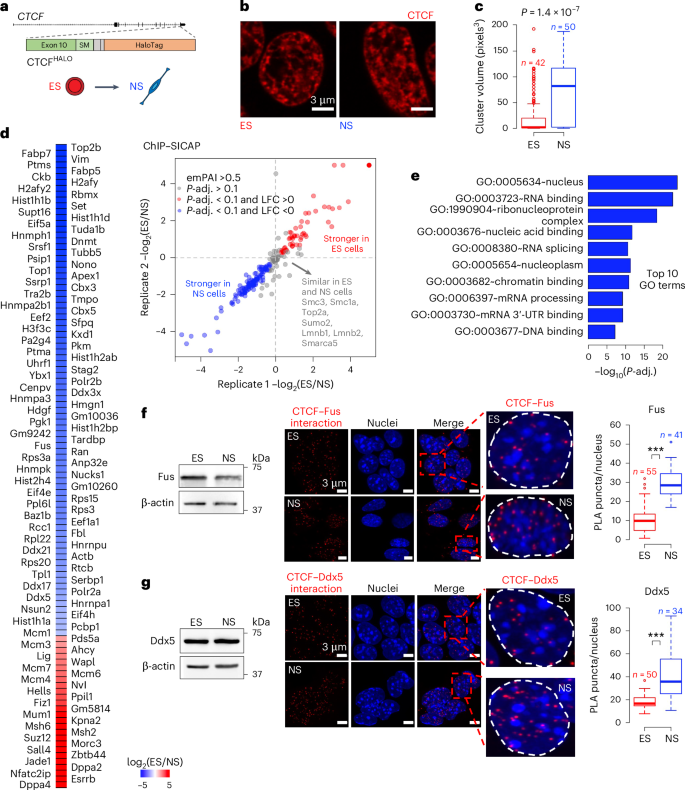
RNA-binding proteins mediate the maturation of chromatin topology during differentiation
Topologically associating domains (TADs) and chromatin architectural loops impact promoter–enhancer interactions, with CCCTC-binding factor (CTCF) defining TAD borders and loop anchors. TAD boundaries and loops progressively strengthen upon embryonic stem (ES) cell differentiation, underscoring the importance of chromatin topology in ontogeny. However, the mechanisms driving this process remain unclear. Here we show a widespread increase in CTCF–RNA-binding protein (RBP) interactions upon ES to neural stem (NS) cell differentiation. While dispensable in ES cells, RBPs reinforce CTCF-anchored chromatin topology in NS cells. We identify Pantr1, a non-coding RNA, as a key facilitator of CTCF–RBP interactions, promoting chromatin maturation. Using acute CTCF degradation, we find that, through its insulator function, CTCF helps maintain neuronal gene silencing in NS cells by acting as a barrier to untimely gene activation during development. Altogether, we reveal a fundamental mechanism driving developmentally linked chromatin structural consolidation and the contribution of this process to the control of gene expression in differentiation. Dehingia, Milewska-Puchała and colleagues find a pervasive lncRNA-mediated increase in interactions between CTCF and RNA-binding proteins during embryonic stem cell differentiation. These interactions reinforce chromatin architecture in neural cells.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


