21种神经和精神疾病的多祖先脑pQTL精细定位和全基因组关联研究整合
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
为了了解脑蛋白表达的共享和祖先特异性遗传控制及其对疾病的影响,我们在来自非洲裔美国人、西班牙裔/拉丁美洲人和非西班牙裔白人供体的1,362个脑蛋白质组中绘制了蛋白质数量性状位点(pqtl)。在多祖先精细定位MESuSiE确定为特定人群中假定的因果pqtl的pqtl中,大多数在三个研究人群中共享,被称为多祖先因果pqtl。这些多祖先因果pqtl在外显子和启动子区域富集。为了研究它们对疾病的影响,我们利用孟德尔随机化和神经和精神疾病全基因组关联研究结果,将858个多祖先因果pqtl作为工具变量建模(欧洲血统参与者有21个特征,非洲血统参与者有10个特征,西班牙裔参与者有4个特征)。我们鉴定出119对多祖先pqtl蛋白对与这些疾病的因果作用一致。值得注意的是,这些多祖先pqtl中有29%是编码变体。这些结果为创建神经和精神疾病的新分子模型奠定了重要的基础,这些模型可能与不同遗传祖先的个体有关。本文章由计算机程序翻译,如有差异,请以英文原文为准。
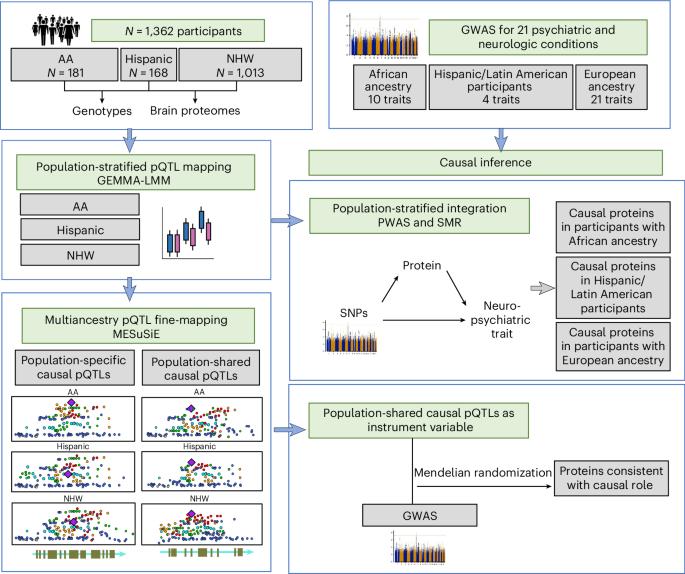
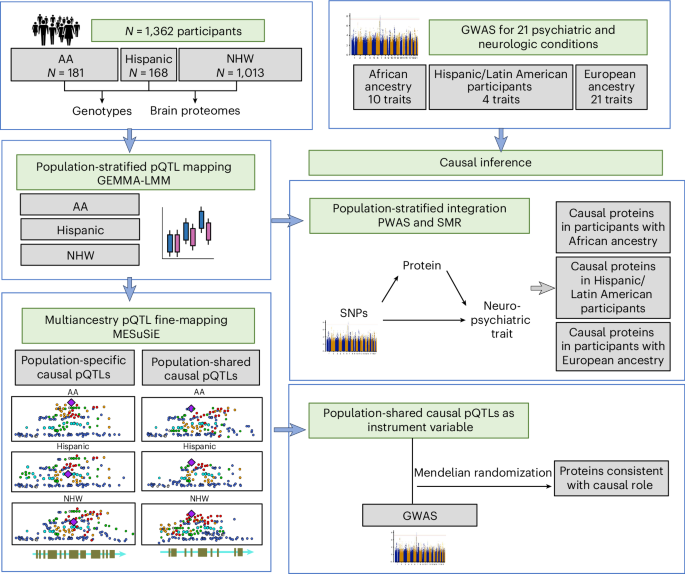
Multiancestry brain pQTL fine-mapping and integration with genome-wide association studies of 21 neurologic and psychiatric conditions
To understand shared and ancestry-specific genetic control of brain protein expression and its ramifications for disease, we mapped protein quantitative trait loci (pQTLs) in 1,362 brain proteomes from African American, Hispanic/Latin American and non-Hispanic white donors. Among the pQTLs that multiancestry fine-mapping MESuSiE confidently assigned as putative causal pQTLs in a specific population, most were shared across the three studied populations and are referred to as multiancestry causal pQTLs. These multiancestry causal pQTLs were enriched for exonic and promoter regions. To investigate their effects on disease, we modeled the 858 multiancestry causal pQTLs as instrumental variables using Mendelian randomization and genome-wide association study results for neurologic and psychiatric conditions (21 traits in participants with European ancestry, 10 in those with African ancestry and 4 in Hispanic participants). We identified 119 multiancestry pQTL–protein pairs consistent with a causal role in these conditions. Remarkably, 29% of the multiancestry pQTLs in these pairs were coding variants. These results lay an important foundation for the creation of new molecular models of neurologic and psychiatric conditions that are likely to be relevant to individuals across different genetic ancestries. Multiancestry fine-mapping of brain protein quantitative trait loci coupled with Mendelian randomization analyses identifies protein–trait pairs consistent with causal effects across neurological and psychiatric conditions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


