PreRBP:基于注意机制的rna -蛋白结合位点预测的可解释深度学习。
IF 2.5
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
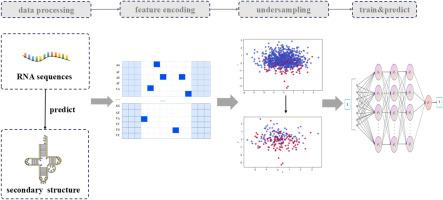
在复杂的基因表达和调控过程中,RNA结合蛋白对RNA起着举足轻重的作用。准确预测rna -蛋白结合位点有助于研究人员更好地了解rna -蛋白结合及其相关机制。而基于机器学习算法的预测技术在识别这些结合位点方面既经济又有效。然而,目前可用的机器学习方法存在一些不足,例如模型的输入特征只考虑RNA序列特征,大多数数据集存在类不平衡。为了解决这些问题,本研究首先使用公开可用的27个rna -蛋白结合位点数据集构建基准数据集。然后,我们使用RNAshapes和EDeN来获得RNA的二级结构。采用高阶编码方法提取隐藏在RNA序列和结构中的关键信息。为了解决数据集中存在的类不平衡问题,本研究利用随机欠采样、NearMiss、ENN和片面选择四种欠采样算法去除负样本中的冗余样本,最后基于卷积神经网络、双向长短期记忆网络构建PreRBP模型预测rna -蛋白结合位点。实验结果表明,本研究使用的模型的平均AUC为0.88,高于现有的其他rna -蛋白结合位点预测方法。此外,为了便于预测,本研究还开发了一种在线预测器。预测器和实验代码可在https://github.com/B12-Comet/RBPPrediction上获得。本文章由计算机程序翻译,如有差异,请以英文原文为准。
PreRBP: Interpretable deep learning for RNA-protein binding site prediction with attention mechanism
In the complex process of gene expression and regulation, RNA-binding proteins occupy a pivotal position for RNA. Accurate prediction of RNA-protein binding sites can help researchers better understand RNA-binding proteins and their related mechanisms. And prediction techniques based on machine learning algorithms are both cost-effective and efficient in identifying these binding sites. However, there are some shortcomings in the currently available machine learning methods, such as the input features of the model only consider RNA sequence features, and most of the datasets suffer from class imbalance. To address these issues, this study first uses the publicly available 27 RNA-protein binding site datasets to construct a benchmark dataset. Then, we use RNAshapes and EDeN to obtain the secondary structure of RNA. Higher-order encoding method is used to extract the key information hidden in the RNA sequences and structures. In order to solve the class imbalance problem existing in the dataset, this study utilizes four undersampling algorithms, namely, random undersampling, NearMiss, ENN, and one-sided selection, to remove redundant samples in the negative samples, and lastly, based on Convolutional Neural Network, Bidirectional Long and Short Term Memory Network, this study constructs model PreRBP to predict RNA-protein binding sites.
The experimental results show that the model used in this study has an average AUC of 0.88, which is higher than other existing RNA-protein binding site prediction methods. Also, for the convenience of prediction, an online predictor is developed in this study. The predictor and experimental codes are available at https://github.com/B12-Comet/RBPPrediction.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Analytical biochemistry
生物-分析化学
CiteScore
5.70
自引率
0.00%
发文量
283
审稿时长
44 days
期刊介绍:
The journal''s title Analytical Biochemistry: Methods in the Biological Sciences declares its broad scope: methods for the basic biological sciences that include biochemistry, molecular genetics, cell biology, proteomics, immunology, bioinformatics and wherever the frontiers of research take the field.
The emphasis is on methods from the strictly analytical to the more preparative that would include novel approaches to protein purification as well as improvements in cell and organ culture. The actual techniques are equally inclusive ranging from aptamers to zymology.
The journal has been particularly active in:
-Analytical techniques for biological molecules-
Aptamer selection and utilization-
Biosensors-
Chromatography-
Cloning, sequencing and mutagenesis-
Electrochemical methods-
Electrophoresis-
Enzyme characterization methods-
Immunological approaches-
Mass spectrometry of proteins and nucleic acids-
Metabolomics-
Nano level techniques-
Optical spectroscopy in all its forms.
The journal is reluctant to include most drug and strictly clinical studies as there are more suitable publication platforms for these types of papers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


