钾通道Kcna5的过度表达改变了斑马鱼再生鳍的骨骼模式。
IF 2.1
3区 生物学
Q2 DEVELOPMENTAL BIOLOGY
引用次数: 0
摘要
骨骼模式依赖于一个复杂的分子和遗传调控网络。然而,我们对控制关节位置和形态发生的途径的理解仍然不完整。先前的研究表明,位于中间位置的Cx43介导的间隙连接细胞间通讯(GJIC)抑制相邻侧骨前体细胞的关节形成,从而决定了骨胳鱼再生鳍中的骨骼模式。在这里,我们测试了Cx43-GJIC通过传播膜电位变化调节关节形成的模型(ΔVm)。为了提供ΔVm足以影响关节形成的证据,我们构建了一个转基因系,表达X. laevis电压门控通道,激振子相关亚家族,温度诱导热休克蛋白70 (hsp70)启动子后面的成员5 (kcna5)。使用这一行,我们证明了xml -kcna5过表达延迟了evx1的表达并导致更长的片段。此外,由于Xl-Kcna5过表达而增加的片段长度需要Cx43。这些发现支持了一个模型,即钾通道与间隙连接通道一起作用,影响斑马鱼再生鳍中的关节形成,从而影响骨骼图案。本文章由计算机程序翻译,如有差异,请以英文原文为准。
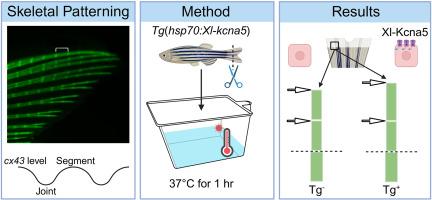
Overexpression of potassium channel Kcna5 alters skeletal patterning in the zebrafish regenerating fin
Skeletal patterning relies on a complex network of molecular and genetic regulators. However, our understanding of pathways governing joint placement and morphogenesis remains incomplete. Prior studies have demonstrated that medially located Cx43 mediated gap junctional intercellular communication (GJIC) inhibits joint formation by the adjacent lateral skeletal precursor cells, and thereby determines skeletal patterning in the teleost regenerating fin. Here, we test the model that Cx43-GJIC regulates joint formation by propagating changes in membrane potential (ΔVm). To provide evidence that ΔVm is sufficient to influence joint formation, we generated a transgenic line that expresses the X. laevis voltage-gated channel, shaker-related subfamily, member 5 (kcna5) behind the temperature-inducible heat shock protein 70 (hsp70) promoter. Using this line, we demonstrate that Xl-kcna5 overexpression delays evx1 expression and causes longer segments. Moreover, the increased segment length in response to Xl-Kcna5 overexpression requires Cx43. These findings support a model whereby potassium channels act together with gap junction channels to influence joint formation, and therefore skeletal patterning, in the zebrafish regenerating fin.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Developmental biology
生物-发育生物学
CiteScore
5.30
自引率
3.70%
发文量
182
审稿时长
1.5 months
期刊介绍:
Developmental Biology (DB) publishes original research on mechanisms of development, differentiation, and growth in animals and plants at the molecular, cellular, genetic and evolutionary levels. Areas of particular emphasis include transcriptional control mechanisms, embryonic patterning, cell-cell interactions, growth factors and signal transduction, and regulatory hierarchies in developing plants and animals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


