人GABA转运体的底物和抑制剂结合3
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
GABA (g-氨基丁酸)转运蛋白3 (GAT3)主要存在于神经胶质细胞中,并通过介导GABA摄取来调节中枢神经系统中GABA的稳态。因此,GAT3已成为治疗癫痫的重要治疗靶点。在这项研究中,我们展示了GAT3在无底物状态下与其底物GABA(选择性抑制剂SNAP-5114)结合的低温电镜(cryo-EM)结构。GAT3以内向的构象结合GABA,而SNAP-5114占据GABA结合口袋,并通过与周围残基的广泛相互作用而稳定。功能研究表明,E66在决定SNAP-5114结合底物模式和特异性方面起着关键作用。综上所述,我们的研究阐明了GAT3的GABA结合机制,揭示了SNAP-5114特异性抑制的分子基础,为开发GAT3亚型选择性抑制剂提供了有价值的见解,这些抑制剂具有治疗癫痫的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
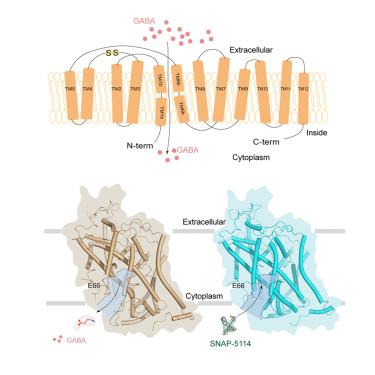
Substrate and inhibitor binding of human GABA transporter 3
GABA (g-aminobutyric acid) transporter 3 (GAT3) is primarily found in glial cells and is essential for regulating GABA homeostasis in the central nervous system by mediating GABA uptake. Consequently, GAT3 has emerged as a significant therapeutic target for the treatment of epilepsy. In this study, we present the cryoelectron microscopy (cryo-EM) structures of GAT3 bound to its substrate GABA, the selective inhibitor SNAP-5114, and in the substrate-free state. GAT3 binds to GABA in an inward-facing conformation, while SNAP-5114 occupies the GABA-binding pocket and is stabilized by extensive interactions with surrounding residues. Functional studies reveal that E66 plays a pivotal role in determining the substrate-binding mode and specificity of SNAP-5114 binding. Taken together, our study clarifies the GABA binding mechanism of GAT3 and reveals the molecular basis for the specific inhibition of SNAP-5114, offering valuable insights for developing GAT3 subtypes selective inhibitors, which hold potential as a treatment for epilepsy.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


