DeepMVP:经过高质量数据训练的深度学习模型可以准确预测PTM位点和变异引起的变化。
IF 32.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
翻译后修饰(PTMs)是蛋白质功能的关键调节因子,其破坏是错义变异导致疾病的关键机制。使用深度学习进行准确的PTM位点预测可以帮助识别PTM改变的变体,但由于缺乏大型、高质量的训练数据集,进展受到限制。在这里,我们介绍了PTMAtlas,一个通过系统再处理241个公共质谱数据集生成的397,524个PTM位点的精选纲要,以及DeepMVP,一个基于PTMAtlas训练的深度学习框架,用于预测PTM位点的磷酸化、乙酰化、甲基化、sumoylation、泛素化和n -糖基化。DeepMVP在所有六种PTM类型中都大大优于现有工具。它在预测ptm改变错义变异方面的应用显示了与实验结果的强烈一致性,并使用文献整理的变异和癌症蛋白质基因组数据集进行了验证。PTMAtlas和DeepMVP共同为PTM研究提供了一个强大的平台,并为通过PTM来评估编码变体的功能后果提供了一个可扩展的框架。本文章由计算机程序翻译,如有差异,请以英文原文为准。
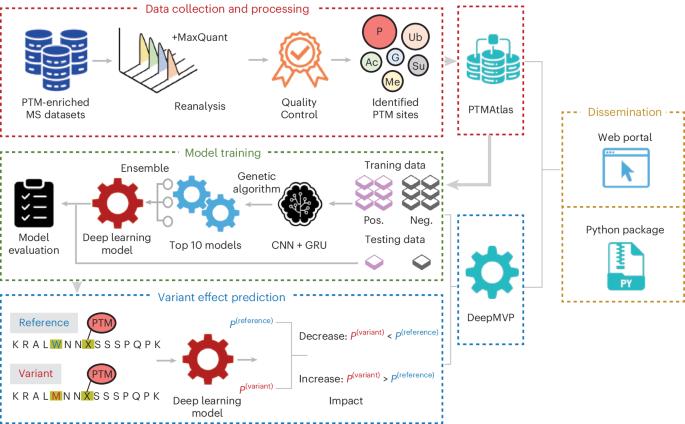
DeepMVP: deep learning models trained on high-quality data accurately predict PTM sites and variant-induced alterations
Post-translational modifications (PTMs) are critical regulators of protein function, and their disruption is a key mechanism by which missense variants contribute to disease. Accurate PTM site prediction using deep learning can help identify PTM-altering variants, but progress has been limited by the lack of large, high-quality training datasets. Here, we introduce PTMAtlas, a curated compendium of 397,524 PTM sites generated through systematic reprocessing of 241 public mass-spectrometry datasets, and DeepMVP, a deep learning framework trained on PTMAtlas to predict PTM sites for phosphorylation, acetylation, methylation, sumoylation, ubiquitination and N-glycosylation. DeepMVP substantially outperforms existing tools across all six PTM types. Its application to predicting PTM-altering missense variants shows strong concordance with experimental results, validated using literature-curated variants and cancer proteogenomic datasets. Together, PTMAtlas and DeepMVP provide a robust platform for PTM research and a scalable framework for assessing the functional consequences of coding variants through the lens of PTMs. DeepMVP is a deep learning framework for predicting PTM sites and variant-induced alterations across six modification types, including phosphorylation, acetylation, methylation, sumoylation, ubiquitination and N-glycosylation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


