龙葵科多花栀子(Gardneria multiflora Makino 1901)叶绿体全基因组特征及系统发育分析。
IF 0.7
4区 生物学
Q4 GENETICS & HEREDITY
Mitochondrial DNA. Part B, Resources
Pub Date : 2025-08-21
eCollection Date: 2025-01-01
DOI:10.1080/23802359.2025.2547917
引用次数: 0
摘要
多花栀子是中国特有的重要药用植物,其根和叶传统上用于治疗目的。在这项研究中,我们报道了多菌根的完整叶绿体基因组。144,920 bp的基因组(GC 37.82%)具有典型的圆形四分体结构:98,036 bp的LSC, 18,382 bp的SSC和两个14,251 bp的IRs。它有125个独特的基因,包括82个蛋白质编码基因,35个tRNA基因和8个rRNA基因。系统发育分析表明,多花田葵与卵田葵亲缘关系较近。本研究提供的数据可以指导今后对该物种进化史的研究。本文章由计算机程序翻译,如有差异,请以英文原文为准。

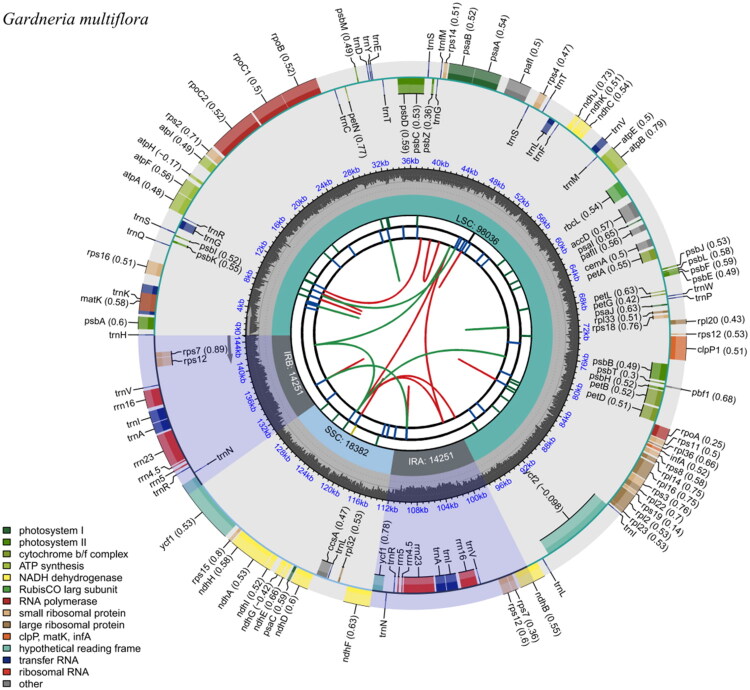
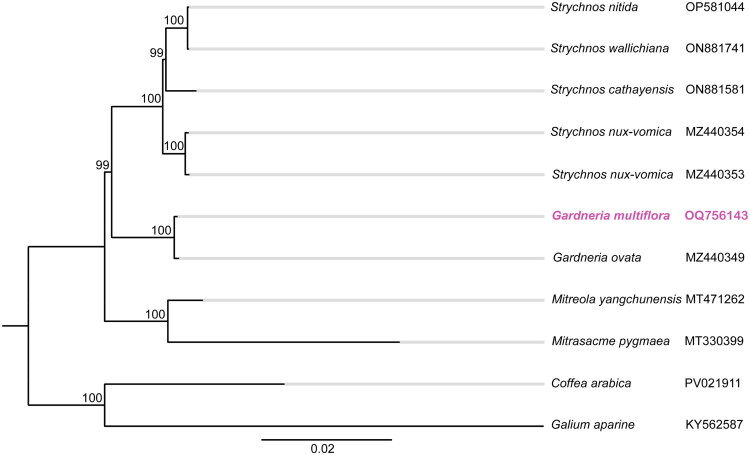
Characteristics and phylogenetic analysis of the complete chloroplast genome of Gardneria multiflora Makino 1901 from the family Loganiaceae.
Gardneria multiflora Makino is an important medicinal plant endemic to China, with its roots and leaves traditionally used for therapeutic purposes. In this study, we report the complete chloroplast genome of G. multiflora. The 144,920 bp genome (GC 37.82%) has a typical circular quadripartite structure: 98,036 bp LSC, 18,382 bp SSC, and two 14,251 bp IRs. It has 125 unique genes, comprising 82 protein-coding, 35 tRNA, and 8 rRNA genes. Phylogenetic analysis revealed that G. multiflora is closely related to G. ovata. The data presented in this study can guide future research on the evolutionary history of this species.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Mitochondrial DNA. Part B, Resources
Biochemistry, Genetics and Molecular Biology-Molecular Biology
CiteScore
1.30
自引率
20.00%
发文量
670
期刊介绍:
This open access journal publishes high-quality and concise research articles reporting the sequence of full mitochondrial genomes, and short communications focusing on the physical, chemical, and biochemical aspects of mtDNA and proteins involved in mtDNA metabolism and interactions.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


