真菌动力蛋白-8马达通过折叠其近端尾部结构域成紧凑的螺旋束进行二聚化
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
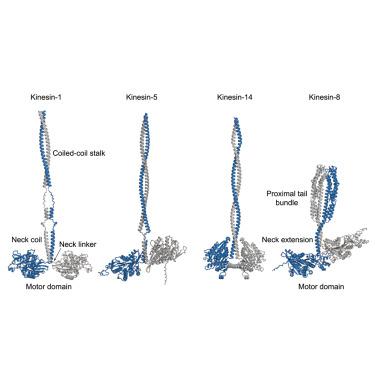
Kinesin-8电机调节着丝微管动力学和控制主轴长度和定位。某些同工异构体通过穿过微管,在正端积累,并解聚末端αβ-微管蛋白亚基来实现这一目标。虽然kinesin-8的马达结构域已被很好地表征,但尾部结构域却鲜为人知。利用白色念珠菌Kip3蛋白作为真菌激酶-8的模型,我们展示了其运动-近端尾部的x射线晶体结构和流体动力学分析,揭示了其在运动二聚化中的作用。这一节段形成一个紧凑的,92 Å-long四螺旋束,而不是在大多数运动蛋白中看到的一个细长的卷曲卷曲的茎。束的稳定主要是通过螺旋1和螺旋3之间的相互作用,以及螺旋2和螺旋4的额外支持。一个灵活的铰链将束分成两个小叶,赋予机械柔韧性和不对称的外部表面。这些独特的功能可以促进与调节元件的相互作用或有助于kinesin-8电机的功能通用性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Fungal kinesin-8 motors dimerize by folding their proximal tail domain into a compact helical bundle
Kinesin-8 motors regulate kinetochore-microtubule dynamics and control spindle length and positioning. Certain isoforms achieve this by traversing microtubules, accumulating at plus-ends, and depolymerizing terminal αβ-tubulin subunits. While the kinesin-8 motor domain is well characterized, the tail domain regions are less understood. Using the Candida albicans Kip3 protein as a model for fungal kinesin-8, we present an X-ray crystal structure and hydrodynamic analysis of its motor-proximal tail segment, revealing its role in motor dimerization. This segment forms a compact, 92 Å-long four-helix bundle, rather than an elongated coiled-coil stalk seen in most kinesins. The bundle is stabilized primarily by interactions between helices one and three, with additional support from helices two and four. A flexible hinge bisects the bundle into two lobules, imparting mechanical pliability and asymmetric exterior surfaces. These unique features may facilitate interactions with regulatory elements or contribute to the functional versatility of kinesin-8 motors.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


