用mRNA显示测量超过200,000种酶促底物的kcat/KM值
IF 19.6
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
摘要
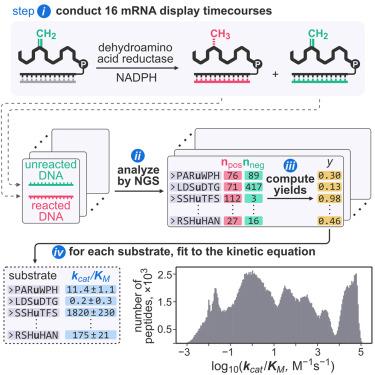
酶学的最新进展是由高通量动力学测量方法实现的。常见的基于仪器的技术可以并行检查数千个酶促反应,但进一步扩展吞吐量可能具有挑战性。在这里,我们建立了DOMEK(基于mRNA显示的酶动力学一次性测量),这是一个集成的实验和计算管道,用于利用mRNA显示衍生的下一代测序数据进行超高通量动力学测量。该方法能准确测定翻译后修饰酶底物的kcat/KM特异性常数。我们通过测量脱氢丙氨酸还原酶的~ 2.86 × 105肽底物的kcat/KM值对平台进行基准测试,并利用所得数据构建底物适应度景观的可解释模型。由此产生的模型准确地将肽底物的反应活化能分解为单个氨基酸的能量贡献,以揭示酶催化的微观和宏观方面。我们的结果建立了一个可推广的,酶不可知的框架缩放动力学测量数以百万计的反应。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Measuring kcat/KM values for over 200,000 enzymatic substrates with mRNA display
Recent advances in enzymology are enabled by the methods for high-throughput kinetic measurements. Common instrumentation-based techniques can examine thousands of enzymatic reactions in parallel, but scaling the throughput further can be challenging. Here, we establish DOMEK (mRNA-display-based one-shot measurement of enzymatic kinetics), an integrated experimental and computational pipeline for ultra-high-throughput kinetic measurements using mRNA display-derived next-generation sequencing data. The method can accurately determine kcat/KM specificity constants of post-translational modification enzyme substrates. We benchmark the platform by measuring kcat/KM values for ∼2.86 × 105 peptide substrates of a dehydroalanine reductase and leverage the resulting data to build interpretable models of the substrate fitness landscape. The resulting model accurately decomposes reaction activation energies of a peptide substrate into energetic contributions of individual amino acids to reveal microscopic and macroscopic aspects of the enzyme catalysis. Our results establish a generalizable, enzyme-agnostic framework for scaling kinetic measurements to millions of reactions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Chem
Environmental Science-Environmental Chemistry
CiteScore
32.40
自引率
1.30%
发文量
281
期刊介绍:
Chem, affiliated with Cell as its sister journal, serves as a platform for groundbreaking research and illustrates how fundamental inquiries in chemistry and its related fields can contribute to addressing future global challenges. It was established in 2016, and is currently edited by Robert Eagling.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


