核梭杆菌OMP生物发生机制的表征
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
具核梭菌是一种革兰氏阴性菌,可引起口腔感染,并与结直肠癌有关。致病性依赖于一种称为自转运蛋白的β桶外膜蛋白(OMP)。omp的生物发生通常由桶状组装机械(BAM)复合物介导。在这项研究中,我们研究了核仁F. OMP生物发生机制的进化、组成和结构。我们的生物信息学和蛋白质组学分析表明,F. nucleatum的OMP生物发生仅由核心成分BamA介导。FnBamA的结构具有明显的特点,包括四个POTRA结构域和一个c端16链β-桶结构域,为倒置二聚体。FnBamA代表了组装机制的原始组成,导致BamA和TamA的复制事件发生在其他谱系(包括变形菌门)从梭杆菌门分裂出来之后。因此,FnBamA可能在所有omp的生物发生中起单一作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
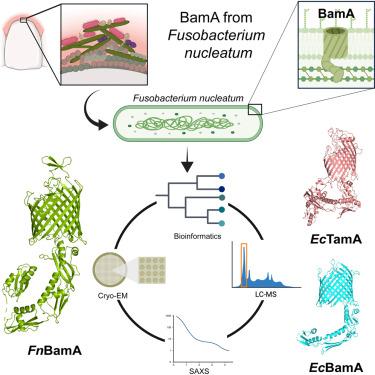
Characterization of the OMP biogenesis machinery in Fusobacterium nucleatum
F. nucleatum is a Gram-negative bacteria that causes oral infections and is linked to colorectal cancer. Pathogenicity relies on a type of β-barrel outer membrane protein (OMP) called an autotransporter. The biogenesis of OMPs is typically mediated by the barrel assembly machinery (BAM) complex. In this study, we investigate the evolution, composition, and structure of the OMP biogenesis machinery in F. nucleatum. Our bioinformatics and proteomics analyses indicate that OMP biogenesis in F. nucleatum is mediated solely by the core component BamA. The structure of FnBamA highlights distinct features, including four POTRA domains and a C-terminal 16-stranded β-barrel domain observed as an inverted dimer. FnBamA represents the original composition of the assembly machinery, and a duplication event that resulted in BamA and TamA occurred after the split of other lineages, including the Proteobacteria, from the Fusobacteria. FnBamA, therefore, likely serves a singular role in the biogenesis of all OMPs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


