系统生物学方法揭示hpv相关头颈部鳞状细胞癌中cnv驱动的lncRNA调控网络
IF 0.7
Q4 GENETICS & HEREDITY
引用次数: 0
摘要
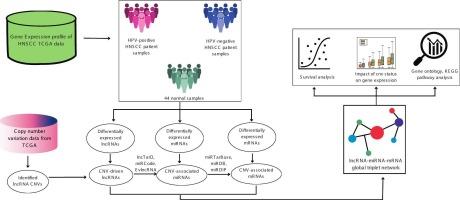
拷贝数变异(CNVs)已被确定为头颈部鳞状细胞癌(HNSCC)发病机制的关键因素。多组学分析提供了对其潜在遗传复杂性的全面理解。因此,本研究旨在研究cnv驱动的长链非编码rna (lncRNAs)对hpv阳性和hpv阴性HNSCC亚型中全球调控三重网络(lncRNA-miRNA-mRNA)的影响。使用TCGA-HNSCC数据鉴定mirna、mrna和cnv相关lncrna的差异表达。随后,目标预测分析能够构建cnv驱动的针对HPV状态的全球三联体网络。基因本体(GO)分析显示,hpv阳性HNSCC中的mrna在癌症相关过程(如细胞增殖和细胞外基质(ECM)组织)中富集。相反,那些hpv阴性的HNSCC主要富集于组织重塑、发育和癌症进展。KEGG通路的富集进一步支持了这些发现。lncRNA MCCC1-AS1的CNV与表达的关系显示,在hpv阳性的HNSCC中,两者之间没有相关性,而在hpv阴性的HNSCC中,lncRNA MCCC1-AS1的基因表达与CNV状态相关。生存分析显示,MCCC1-AS1拷贝数增加的患者与较短的生存时间相关,提示其作为预后生物标志物的潜力。这些发现强调了cnv驱动的lncrna在HNSCC分子格局中的重要性,并提示MCCC1-AS1可能作为一个有希望的靶点,在诊断和治疗策略方面进行进一步的研究。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A systems biology approach to uncover CNV-driven lncRNA regulatory networks in HPV-associated head and neck squamous cell carcinoma
Copy number variations (CNVs) have been identified as critical contributors to head and neck squamous cell carcinoma (HNSCC) pathogenesis. Multi-omics analyses offer a comprehensive understanding of its underlying genetic complexities. Therefore, this study aims to examine the CNV-driven long non-coding RNAs (lncRNAs) influencing global regulatory triplet networks (lncRNA–miRNA–mRNA) in HPV-positive and HPV-negative HNSCC subtypes. Differential expression of miRNAs, mRNAs, and CNV-associated lncRNAs were identified using TCGA-HNSCC data. Subsequently, target prediction analyses enabled the construction of CNV-driven global triplet networks specific to HPV status. Gene Ontology (GO) analysis revealed that mRNAs in HPV-positive HNSCC were enriched in cancer-associated processes such as cell proliferation and extracellular matrix (ECM) organization. In contrast, those in HPV-negative HNSCC were primarily enriched in tissue remodeling, development, and cancer progression. KEGG pathway enrichment further supported these findings. The relationship between the CNV of lncRNA MCCC1-AS1 and its expression has revealed that there was no correlation between them in the HPV-positive HNSCC, while in the HPV-negative HNSCC, the gene expression of lncRNA MCCC1-AS1 was correlated with the CNV status. Survival analysis disclosed that the patient with a copy number gain of MCCC1-AS1 was associated with a shorter survival time, suggesting its potential as a prognostic biomarker. These findings highlight the significance of CNV-driven lncRNAs in the molecular landscape of HNSCC and suggest that MCCC1-AS1 may serve as a promising target for further investigation in diagnostic and therapeutic strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Human Gene
Biochemistry, Genetics and Molecular Biology (General), Genetics
CiteScore
1.60
自引率
0.00%
发文量
0
审稿时长
54 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


