Mettl15-Mettl17调节从早期到晚期前线粒体的转变
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
线粒体小亚基的生物发生涉及一个动态的装配因子网络。保守的甲基转移酶Mettl15和Mettl17作用于rRNA的溶剂暴露表面。Mettl17的结合与早期组装阶段有关,而Mettl15则参与后期组装阶段。在这里,我们将布鲁氏锥虫的结构数据与哺乳动物同源物和分子动力学模拟相结合。我们揭示了Mettl15和Mettl17在中间步骤中的相互作用如何将小亚基组装的不同阶段联系起来。分析提出了一种模式,其中Mettl17作为Mettl15招聘的平台。随后发布的Mettl17允许改变Mettl15的构象以识别底物。甲基化后,Mettl15采用松散结合状态,导致其被起始因子取代,完成组装。综上所述,我们的研究结果表明,组装因子Mettl15和Mettl17共同调控了生物发生过程。本文章由计算机程序翻译,如有差异,请以英文原文为准。
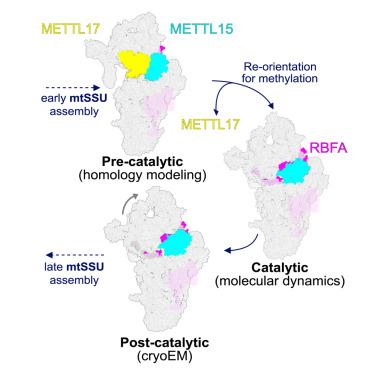
Mettl15-Mettl17 modulates the transition from early to late pre-mitoribosome
The biogenesis of the mitoribosomal small subunit involves a dynamic network of assembly factors. Conserved methyltransferases Mettl15 and Mettl17 act on the solvent-exposed surface of rRNA. Binding of Mettl17 is associated with the early assembly stage, whereas Mettl15 is involved in the late stage. Here, we integrate structural data from Trypanosoma brucei with mammalian homologs and molecular dynamics simulations. We reveal how the interplay of Mettl15 and Mettl17 in intermediate steps links the distinct stages of small subunit assembly. The analysis suggests a model wherein Mettl17 acts as a platform for Mettl15 recruitment. Subsequent release of Mettl17 allows a conformational change of Mettl15 for substrate recognition. Upon methylation, Mettl15 adopts a loosely bound state which leads to its replacement by initiation factors, concluding the assembly. Together, our results indicate that assembly factors Mettl15 and Mettl17 cooperate to regulate the biogenesis process.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


