果蝇隐花色素的光诱导构象开关和磁敏感性
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
隐色素是一种具有多种生物学作用的光敏黄蛋白,其中包括一种被提出的磁接受功能。这种机制依赖于黄素发色团与蛋白质支架内的一条色氨酸残基链的磁敏光化学反应。然而,蛋白质介导的磁信号转导机制尚不清楚。我们通过氢-氘交换质谱,辅以分子动力学模拟和腔衰荡光谱,研究了一种原型隐花色素DmCRY对光化学激活的响应。我们能够以接近残基水平的分辨率测量DmCRY的动力学,揭示了蛋白质c端尾部可逆的、长寿命的蓝光诱导的构象变化。这种假定的信号状态在不同的光照条件下得到了验证,并通过检查电子传递链受到干扰的DmCRY变体。我们的研究结果显示了黄素发色团的光化学行为如何产生DmCRY状态,这可能会引发下游相互作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
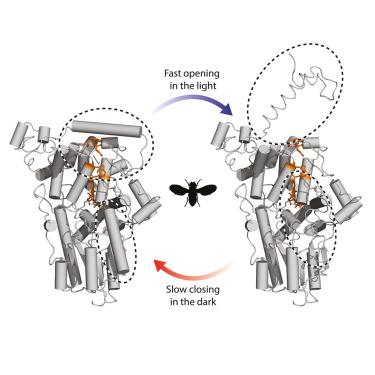
Light-induced conformational switching and magnetic sensitivity of Drosophila cryptochrome
Cryptochromes are light-sensitive flavoproteins with various biological roles, including a proposed function in magnetoreception. This mechanism rests on a magnetically sensitive photochemical reaction of the flavin chromophore with a chain of tryptophan residues within the protein scaffold. However, the protein-mediated mechanisms of magnetic signal transduction are unclear. We have examined the response of an archetypal cryptochrome, DmCRY, to photochemical activation by means of hydrogen-deuterium exchange mass spectrometry, complemented by molecular dynamics simulations and cavity ring-down spectroscopy. We were able to measure the dynamics of DmCRY at near-residue level resolution, revealing a reversible, long-lived, blue-light induced conformational change in the protein’s C-terminal tail. This putative signaling state was validated using different illumination conditions, and by examining DmCRY variants in which the electron transfer chain was perturbed. Our results show how the photochemical behavior of the flavin chromophore generates a state of DmCRY that may initiate downstream interactions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


