靶向KDM5C去甲基酶活性的选择性肽抑制剂的设计
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
翻译后修饰,特别是蛋白质赖氨酸去甲基化,复杂地调节着多种细胞过程。这种修饰的失调通常通过干扰底物蛋白的功能、稳定性和相互作用而引发人类疾病。赖氨酸去甲基化酶(kdm),如KDM5家族,在去除甲基标记中至关重要。特别是,KDM5C因其在癌症生物学和耐药中的作用而受到重视。这些酶,专门用于擦除赖氨酸甲基化标记-特别是从组蛋白H3赖氨酸4 (H3K4) -直接影响基因转录。本研究开创了KDM5C去甲基酶活性肽抑制剂的设计。这种新型抑制剂对KDM5C的选择性优于其他家族成员。有趣的是,体内实验表明,这种抑制剂可显著降低肿瘤生长。这些发现突出了靶向KDM5C抑制作为结肠癌治疗策略的潜力。此外,这些发现强调了肽抑制剂作为靶向治疗的前景,强调了它们在改变癌症治疗轨迹方面的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
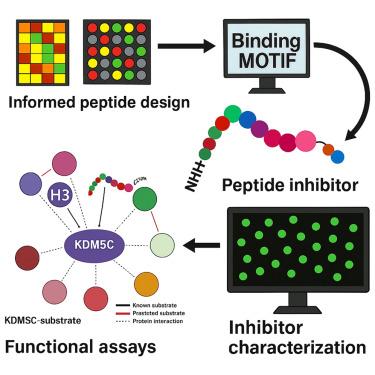
Design of a selective peptide inhibitor targeting KDM5C demethylase activity
Post-translational modifications, particularly protein lysine demethylation, intricately regulate diverse cellular processes. Dysregulation of this modification often precipitates human pathologies by perturbing substrate protein functions, stability, and interactions. Lysine demethylases (KDMs), such as the KDM5 family, are crucial in removing methyl marks. In particular, KDM5C has gained prominence for its role in cancer biology and drug resistance. These enzymes, specializing in erasing lysine methylation marks—especially from histone H3 lysine 4 (H3K4)—directly influence gene transcription. This study pioneers the design of a peptide inhibitor of KDM5C demethylase activity. This novel inhibitor displays remarkable selectivity for KDM5C over other family members. Intriguingly, in vivo experiments demonstrate that this inhibitor significantly reduces tumor growth. These findings highlight the potential of targeting KDM5C inhibition as a strategy for colon cancer treatment. Moreover, these findings underscore the promise of peptide inhibitors as targeted therapies, emphasizing their potential in altering the trajectory of cancer therapeutics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


