基于质谱的代谢组学和脂质组学分析对糖尿病视网膜病变的分期进行分层
IF 7.4
3区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
摘要
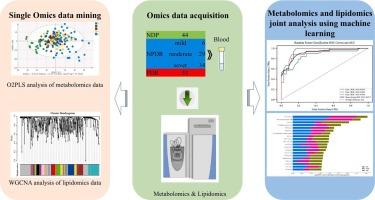
目的:糖尿病视网膜病变是糖尿病的代谢并发症。本研究旨在阐明与糖尿病视网膜病变进展相关的代谢和脂质组学改变,并确定其诊断和分期的生物标志物。方法:我们分析了167例患者的962种分子的血清谱,包括653种脂质和309种极性小代谢物,其中45例无视网膜病变,69例患有不同程度的非增殖性糖尿病视网膜病变,53例患有增殖性糖尿病视网膜病变。统计模型应用于单组学数据集,机器学习算法用于整合代谢组学和脂质组学数据,以识别最能区分疾病分期的特征。结果在非增殖期和增殖期,spine和鞘脂代谢均发生显著变化。非增殖性疾病中酪氨酸代谢被破坏,而增殖性疾病中甘氨酸-丝氨酸-苏氨酸代谢突出。进展与鞘磷脂、磷脂酰胆碱和溶血磷脂酰胆碱减少有关。在10种机器学习模型中,k近邻算法的分类性能最高。脂质ST 24:1;O4和代谢物β -羟基异戊酸是主要贡献者。结论基于质谱技术的代谢组学/脂质组学与机器学习技术相结合,可以准确地对糖尿病视网膜病变进行分层,并发现新的分子标志物,为糖尿病视网膜病变的早期诊断和靶向干预提供支持。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Mass spectrometry-based metabolomic and lipidomic profiling stratifies stages of diabetic retinopathy
Aims
Diabetic retinopathy is a metabolic complication of diabetes. This study aimed to elucidate metabolic and lipidomic alterations associated with diabetic retinopathy progression and identify biomarkers for its diagnosis and staging.
Methods
We analyzed serum profiles of 962 molecules, including 653 lipids and 309 polar small metabolites, from 167 individuals: 45 without retinopathy, 69 with non-proliferative diabetic retinopathy of varying severity, and 53 with proliferative diabetic retinopathy. Statistical models were applied to single-omics datasets, and machine learning algorithms were used to integrate metabolomic and lipidomic data for identifying features that best discriminate disease stages.
Results
Purine and sphingolipid metabolism were significantly altered in both non-proliferative and proliferative stages. Tyrosine metabolism was disrupted in non-proliferative disease, while glycine-serine-threonine metabolism was prominent in proliferative disease. Progression was associated with reduced sphingomyelins, phosphatidylcholines, and lysophosphatidylcholines. Among ten machine learning models, the K-nearest neighbor algorithm achieved the highest classification performance. The lipid ST 24:1;O4 and metabolite beta-hydroxyisovaleric acid were the top contributors.
Conclusions
Mass spectrometry-based metabolomics/lipidomics integrated with machine learning accurately stratify diabetic retinopathy and identify novel molecular markers, and may support early diagnosis and targeted intervention of diabetic retinopathy.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Diabetes research and clinical practice
医学-内分泌学与代谢
CiteScore
10.30
自引率
3.90%
发文量
862
审稿时长
32 days
期刊介绍:
Diabetes Research and Clinical Practice is an international journal for health-care providers and clinically oriented researchers that publishes high-quality original research articles and expert reviews in diabetes and related areas. The role of the journal is to provide a venue for dissemination of knowledge and discussion of topics related to diabetes clinical research and patient care. Topics of focus include translational science, genetics, immunology, nutrition, psychosocial research, epidemiology, prevention, socio-economic research, complications, new treatments, technologies and therapy.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


