小鼠胎儿发育过程中生物钟基因表达的个体发生
IF 2.2
Q3 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
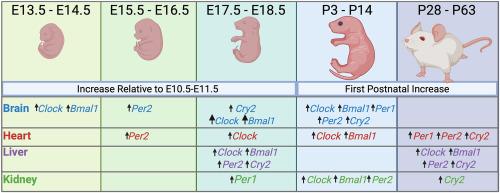
视交叉上核和周围组织中的生物钟在大约24小时的周期内调节关键的生理和细胞系统。对哺乳动物发育过程中生物钟机制的个体发生机制的了解尚不完整。因此,我们使用小鼠作为模型和先前发表的RNAseq数据集来确定在胎儿和出生后的大脑、心脏、肝脏和肾脏中,调节生物钟的核心基因的表达何时增加转录物丰度。在所有组织和时间点检测到所有六个核心基因(Clock, Bmal1, Per1, Per2, Cry1和Cry2)的转录本。对于大脑而言,时钟在E13.5时略有增加,在E18.5时增加较多。同样,Bmal1转录本丰度在E14.5时略有增加,在E17.5时增加的程度更大,Per2转录本在E15.5时略有增加,并保持不变直到出生后,Cry2转录本在E18.5时略有增加。对于肝脏,大多数基因的转录丰度在妊娠结束时增加,Clock在妊娠第18.5期增加,Bmal1、Per2和Cry2在妊娠第17.5期增加。出生前在心脏中表达增加的基因为Clock (E18.5)和Per2 (E15.5),在肾脏中表达增加的基因为Clock (E18.5)、Bmal1 (E15.5)和Per1 (P18.5)。在所有组织中,除Cry1(所有组织)和Per1(肝脏和肾脏)外,大多数基因的转录丰度在出生后进一步增加。结果支持了分子钟组织在胎儿大脑和肝脏中比在胎儿心脏和肾脏中更先进的观点。此外,基于基因表达的变化,分子钟的组成部分在出生后继续成熟。本文章由计算机程序翻译,如有差异,请以英文原文为准。
The ontogeny of circadian clock gene expression during mouse fetal development
The circadian clock in the suprachiasmatic nucleus and peripheral tissues functions to regulate key physiological and cellular systems in a cycle approximating 24 h. Understanding the ontogeny of the circadian clock mechanism during mammalian development is incomplete. Accordingly, we used the mouse as a model and a previously published RNAseq dataset to determine when expression of core genes regulating the circadian clock increase in transcript abundance in fetal and postnatal brain, heart, liver, and kidney. Transcripts for all six core genes examined (Clock, Bmal1, Per1, Per2, Cry1, and Cry2) were identified in all tissues and time points. For brain, there was a small increase in Clock at E13.5 and a larger increase at E18.5. Similarly, Bmal1 transcript abundance increased slightly at E14.5 and to a greater degree at E17.5, Per2 increased slightly at E15.5 and remained constant until after birth and Cry2 increased slightly at E18.5. For liver, transcript abundance of most genes increased at the end of gestation, with increases observed for Clock at E18.5, and for Bmal1, Per2, and Cry2 at E17.5. The genes whose expression increased before birth in heart was Clock (at E18.5), and Per2 (at E15.5) and the genes whose expression increased before birth in kidney were Clock (E18.5), Bmal1 (E15.5) and Per1 (P18.5). For all tissues, there were further increases in transcript abundance for most genes in the postnatal period with the exception of Cry1 (all tissues) and Per1 (liver and kidney). Results support the idea that the organization of the molecular clock is more advanced for the fetal brain and liver than for fetal heart and kidney. Furthermore, based on changes in gene expression, components of the molecular clock continued to mature after birth.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Biochemistry and Biophysics Reports
Biochemistry, Genetics and Molecular Biology-Biophysics
CiteScore
4.60
自引率
0.00%
发文量
191
审稿时长
59 days
期刊介绍:
Open access, online only, peer-reviewed international journal in the Life Sciences, established in 2014 Biochemistry and Biophysics Reports (BB Reports) publishes original research in all aspects of Biochemistry, Biophysics and related areas like Molecular and Cell Biology. BB Reports welcomes solid though more preliminary, descriptive and small scale results if they have the potential to stimulate and/or contribute to future research, leading to new insights or hypothesis. Primary criteria for acceptance is that the work is original, scientifically and technically sound and provides valuable knowledge to life sciences research. We strongly believe all results deserve to be published and documented for the advancement of science. BB Reports specifically appreciates receiving reports on: Negative results, Replication studies, Reanalysis of previous datasets.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


