ergd驱动的前列腺癌起始依赖于细胞背景,需要KMT2A和DOT1L
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
尽管前列腺癌中ERG转录因子易位的发生率很高,但其致瘤性机制仍知之甚少。通过谱系追踪,我们发现ERG的肿瘤启动活性存在于共表达管腔基因(BasalLum)的小鼠基底细胞亚群中,而不是在更大的ERG+管腔细胞群中。在ERG激活后,BasalLum细胞产生具有干细胞样特征的高增殖中间细胞(IM),这些细胞共同表达基底、管腔、丘和俱乐部标记基因,然后转化为Krt8+管腔细胞。ERG+人前列腺癌的转录组学分析证实存在罕见的ERG+ BasalLum细胞,以及与预后较差相关的IM细胞。单细胞分析显示,ERG+ IM细胞的染色质状态丰富,STAT3转录因子结合位点丰富,KMT2A/MLL1和DOT1L的表达升高,这三种基因都是体内ERG驱动的致瘤性所必需的。除了提供翻译机会,这项工作还说明了单细胞方法与谱系追踪相结合如何识别从批量分析中不明显的癌症脆弱性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
ERG-driven prostate cancer initiation is cell-context dependent and requires KMT2A and DOT1L
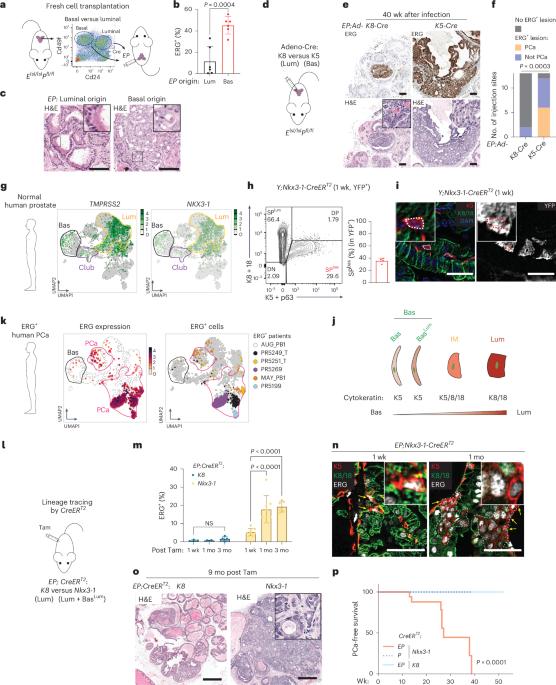
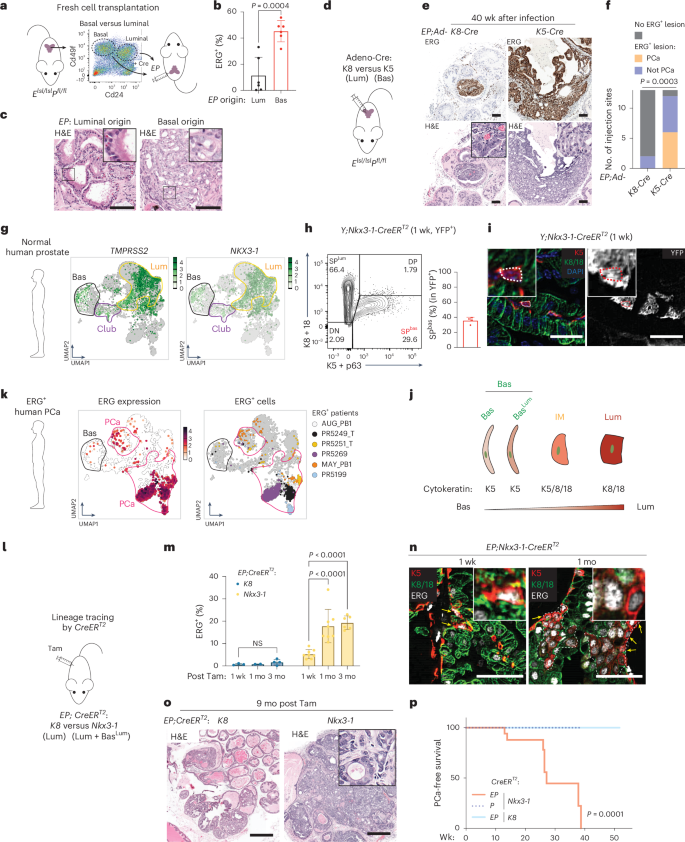
Despite the high prevalence of ERG transcription factor translocations in prostate cancer, the mechanism of tumorigenicity remains poorly understood. Using lineage tracing, we find the tumor-initiating activity of ERG resides in a subpopulation of murine basal cells that coexpress luminal genes (BasalLum) and not in the larger population of ERG+ luminal cells. Upon ERG activation, BasalLum cells give rise to highly proliferative intermediate (IM) cells with stem-like features that coexpress basal, luminal, hillock and club marker genes, before transitioning to Krt8+ luminal cells. Transcriptomic analysis of ERG+ human prostate cancers confirms the presence of rare ERG+ BasalLum cells, as well as IM cells whose presence is associated with a worse prognosis. Single-cell analysis revealed a chromatin state in ERG+ IM cells enriched for STAT3 transcription factor binding sites and elevated expression of the KMT2A/MLL1 and DOT1L, all three of which are essential for ERG-driven tumorigenicity in vivo. In addition to providing translational opportunities, this work illustrates how single-cell approaches combined with lineage tracing can identify cancer vulnerabilities not evident from bulk analysis. Lineage tracing in mice identifies a subpopulation of basal cells that express Tmprss2 and Nkx3 as the origin of ERG-driven prostate cancer. Upon expansion, these cells show an enrichment for STAT3 chromatin binding and elevated expression of KMT2A and DOT1L as dependencies for ERG oncogenicity.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


