ImageDDI:用于药物-药物相互作用预测的图像增强分子基序序列表示
IF 15.5
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
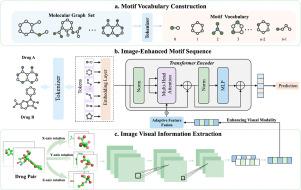
为了减轻同时使用多种药物对健康的潜在不利影响,包括意想不到的副作用和相互作用,准确识别和预测药物-药物相互作用(ddi)被认为是深度学习领域的一项关键任务。尽管现有的方法表现出了良好的性能,但由于ddi基本上是由基序相互作用而不是整体药物结构引起的,因此它们受到基于基序的有限功能表征学习的瓶颈。在本文中,我们提出了一个用于DDI预测的图像增强分子基序序列表示框架,称为ImageDDI,它代表了来自全局和局部结构的一对药物。具体来说,ImageDDI将分子标记为功能基序。为了有效地表示药物对,它们的基序被组合成一个单一的序列,并使用基于变压器的编码器嵌入,从局部结构表示开始。通过利用药物对之间的关联,ImageDDI利用全局分子图像信息(如纹理、阴影、颜色和平面空间关系)进一步增强分子的空间表示。为了将分子视觉信息整合到功能基序序列中,ImageDDI采用了自适应特征融合,通过动态调整特征表示的融合过程,增强了ImageDDI的泛化能力。在广泛使用的数据集上的实验结果表明,ImageDDI优于最先进的方法。此外,大量的实验表明,与其他模型相比,ImageDDI在2D和3D图像增强场景中都取得了具有竞争力的性能。本文章由计算机程序翻译,如有差异,请以英文原文为准。
ImageDDI: Image-enhanced molecular motif sequence representation for drug–drug interaction prediction
To mitigate the potential adverse health effects of simultaneous multi-drug use, including unexpected side effects and interactions, accurately identifying and predicting drug–drug interactions (DDIs) is considered a crucial task in the field of deep learning. Although existing methods have demonstrated promising performance, they suffer from the bottleneck of limited functional motif-based representation learning, as DDIs are fundamentally caused by motif interactions rather than the overall drug structures. In this paper, we propose an Image-enhanced molecular motif sequence representation framework for DDI prediction, called ImageDDI, which represents a pair of drugs from both global and local structures. Specifically, ImageDDI tokenizes molecules into functional motifs. To effectively represent a drug pair, their motifs are combined into a single sequence and embedded using a transformer-based encoder, starting from the local structure representation. By leveraging the associations between drug pairs, ImageDDI further enhances the spatial representation of molecules using global molecular image information (e.g. texture, shadow, color, and planar spatial relationships). To integrate molecular visual information into functional motif sequence, ImageDDI employs Adaptive Feature Fusion, enhancing the generalization of ImageDDI by dynamically adapting the fusion process of feature representations. Experimental results on widely used datasets demonstrate that ImageDDI outperforms state-of-the-art methods. Moreover, extensive experiments show that ImageDDI achieved competitive performance in both 2D and 3D image-enhanced scenarios compared to other models.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Information Fusion
工程技术-计算机:理论方法
CiteScore
33.20
自引率
4.30%
发文量
161
审稿时长
7.9 months
期刊介绍:
Information Fusion serves as a central platform for showcasing advancements in multi-sensor, multi-source, multi-process information fusion, fostering collaboration among diverse disciplines driving its progress. It is the leading outlet for sharing research and development in this field, focusing on architectures, algorithms, and applications. Papers dealing with fundamental theoretical analyses as well as those demonstrating their application to real-world problems will be welcome.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


