新生抗生素设计的生成式深度学习方法
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
抗菌素耐药性危机需要结构不同的抗生素。虽然深度学习方法可以从现有的文库中识别抗菌化合物,但结构的新颖性仍然有限。在这里,我们通过两种方法开发了一个生成式人工智能框架,用于设计新生抗生素:基于片段的方法,全面筛选107个针对淋病奈瑟菌或金黄色葡萄球菌的化学片段,随后扩展有希望的片段,以及不受约束的新生化合物生成,每一种方法都使用遗传算法和变分自编码器。在所合成的24个化合物中,有7个具有选择性抗菌活性。在淋病奈瑟菌阴道感染和耐甲氧西林金黄色葡萄球菌皮肤感染小鼠模型中,两种铅化合物对多重耐药分离株具有不同作用机制的抗菌效果,并减少了体内细菌负荷。我们进一步验证了这两类化合物的结构类似物的抗菌作用。我们的方法使生成式深度学习引导的新抗生素设计成为可能,为绘制化学空间的未知区域提供了一个平台。本文章由计算机程序翻译,如有差异,请以英文原文为准。
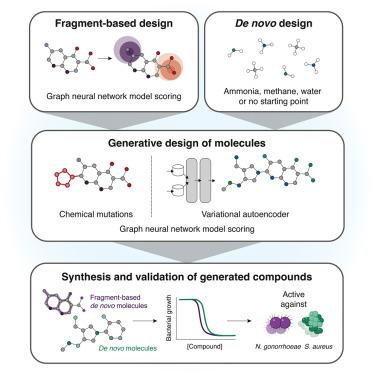
A generative deep learning approach to de novo antibiotic design
The antimicrobial resistance crisis necessitates structurally distinct antibiotics. While deep learning approaches can identify antibacterial compounds from existing libraries, structural novelty remains limited. Here, we developed a generative artificial intelligence framework for designing de novo antibiotics through two approaches: a fragment-based method to comprehensively screen >107 chemical fragments in silico against Neisseria gonorrhoeae or Staphylococcus aureus, subsequently expanding promising fragments, and an unconstrained de novo compound generation, each using genetic algorithms and variational autoencoders. Of 24 synthesized compounds, seven demonstrated selective antibacterial activity. Two lead compounds exhibited bactericidal efficacy against multidrug-resistant isolates with distinct mechanisms of action and reduced bacterial burden in vivo in mouse models of N. gonorrhoeae vaginal infection and methicillin-resistant S. aureus skin infection. We further validated structural analogs for both compound classes as antibacterial. Our approach enables the generative deep-learning-guided design of de novo antibiotics, providing a platform for mapping uncharted regions of chemical space.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


