假单胞菌通过一个单一的调节系统协调对病毒和细菌的先天防御
IF 18.7
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
摘要
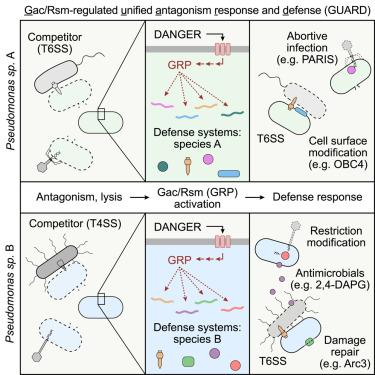
细菌细胞不断受到其他细菌和病毒的生存威胁。对付这些威胁的机制有充分的记录;然而,对这些不同防御要素的调控仍然知之甚少。在这里,我们描述了一种全基因组的、协调的、高效的免疫反应,称为Gac/ rsm调节的统一拮抗反应和防御(GUARD),它通过单一的调节途径保护免受细菌和病毒的威胁。生物信息学分析揭示了铜绿假单胞菌中Gac/Rsm调控途径(GRP)的一种全假单胞菌形式,这是一种已建立的危险感知系统。对不同假单胞菌种类的蛋白质组学研究表明,该途径调节了涉及防御细菌和噬菌体威胁的大量可变因素。专注于P. protegens,我们确定了这些因素对多种形式的细菌拮抗和几种噬菌体的深刻表型后果。总之,我们的研究结果表明,细菌像多细胞真核生物一样,将危险感知与抗菌和抗病毒武器的免疫反应激活结合起来。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Pseudomonads coordinate innate defense against viruses and bacteria with a single regulatory system
Bacterial cells live under constant existential threats imposed by other bacteria and viruses. Mechanisms for contending with these threats are well documented; however, the regulation of these diverse defense elements remains poorly understood. Here, we describe a genome-wide, coordinated, and highly effective immune response, termed Gac/Rsm-regulated unified antagonism response and defense (GUARD), that protects against bacterial and viral threats using a single regulatory pathway. Bioinformatic analyses reveal a Pseudomonas-wide form of the Gac/Rsm regulatory pathway (GRP), an established danger-sensing system in P. aeruginosa. Proteomic studies of diverse Pseudomonas species show that the pathway regulates a large and variable suite of factors implicated in defense against both bacterial and phage threats. Focusing on P. protegens, we identify profound phenotypic consequences of these factors against multiple forms of bacterial antagonism and several phages. Together, our results reveal that bacteria, like multicellular eukaryotes, couple danger sensing to the activation of an immune response with antibacterial and antiviral arms.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


