人类肠道微生物群的亚种为深入的微生物组研究提供了隐含的信息
IF 18.7
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
摘要
由于基因含量的变化,单个物种内的微生物菌株可以表现出不同的功能特征,并且通常表现出个体特异性,这可能会模糊公正的关联并阻碍演绎研究。在这里,我们以一致注释的操作亚种单位(OSU)分辨率以无偏、队列独立的方式全面定义了人类肠道微生物群,表明这种方法可以在保持特异性和提高研究间可重复性的同时推广到不同的全球人群。我们开发了panhashome——一种基于草图的方法,用于快速亚种和物种量化和识别驱动种内变异的基因——并表明亚种携带在物种水平上无法检测到的隐性信息。我们确定了与结直肠癌(CRC)相关的亚种,其兄弟亚种或物种没有,而基于亚种的机器学习CRC诊断算法优于物种水平的方法。这种亚种目录允许识别驱动亚种之间功能差异的基因,作为机制理解微生物组-表型相互作用的基本步骤。本文章由计算机程序翻译,如有差异,请以英文原文为准。
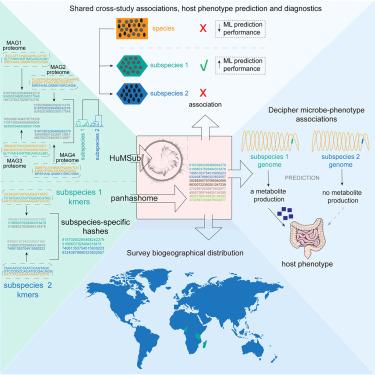
Subspecies of the human gut microbiota carry implicit information for in-depth microbiome research
Microbial strains within a single species can exhibit distinct functional characteristics due to variations in gene content and often show individual specificity, which can obscure unbiased associations and hinder deductive research. Here, we comprehensively define the human gut microbiota at a consistently annotated operational subspecies unit (OSU) resolution in an unbiased, cohort-independent manner, demonstrating that this approach can generalize across diverse global populations while maintaining specificity and improving interstudy reproducibility. We develop panhashome—a sketching-based method for rapid subspecies and species quantification and identification of genes that drive intraspecies variations—and show that subspecies carry implicit information undetectable at the species level. We identify subspecies associated with colorectal cancer (CRC) whose sibling subspecies or species are not, while a machine-learning CRC diagnostic algorithm based on subspecies outperformed species-level methods. This subspecies catalog allows identification of genes that drive functional differences between subspecies as a fundamental step in mechanistically understanding microbiome-phenotype interactions.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


