泛基因组背景揭示了种内植物NLR进化的程度
IF 18.7
1区 医学
Q1 MICROBIOLOGY
引用次数: 0
摘要
核苷酸结合丰富亮氨酸重复序列(NLR)蛋白是植物免疫系统的主要组成部分,识别病原体效应并引发防御反应。由于病原体效应谱的多样性,nlr具有非凡的序列、结构和调控变异性。虽然对NLR多样性的贡献过程已经被确定,但NLR在其基因组背景下以及沿着多样性的多个轴的精确进化一直难以追踪。我们整合了基因组特异性的全长转录本、同源性和转座元件信息,对17个不同拟南芥材料中的3789个NLRs进行了注释。我们定义了121个泛基因组NLR邻域,它们在大小、内容和复杂性方面差异很大。NLR在许多轴上是不同的,并且需要多个度量来完全捕获NLR的变化。基于这些发现,我们提出多样性产生的多样性是维持植物功能性“适应性”免疫系统的基础,机制研究应考虑免疫系统多样性的多个轴。本文章由计算机程序翻译,如有差异,请以英文原文为准。
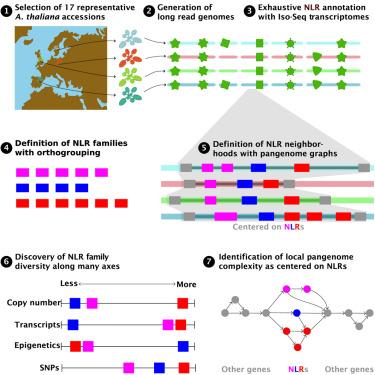
Pangenomic context reveals the extent of intraspecific plant NLR evolution
Nucleotide-binding leucine-rich repeat (NLR) proteins are major components of the plant immune system, recognizing pathogen effectors and triggering defense responses. Because of the diversity of pathogen effector repertoires, NLRs have extraordinary sequence, structural, and regulatory variability. Although processes contributing to NLR diversity have been identified, the precise evolution of NLRs in their genomic context and along the multiple axes of diversity has been difficult to trace. We integrate genome-specific full-length transcript, homology, and transposable element information to annotate 3,789 NLRs in 17 diverse Arabidopsis thaliana accessions. We define 121 pangenomic NLR neighborhoods, which vary greatly in size, content, and complexity. NLRs are diverse across many axes, and multiple metrics are required to fully capture NLR variation. Based on these findings, we propose that diversity in diversity generation is fundamental to maintaining a functionally “adaptive” immune system in plants and that mechanistic studies should consider multiple axes of immune system diversity.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell host & microbe
生物-微生物学
CiteScore
45.10
自引率
1.70%
发文量
201
审稿时长
4-8 weeks
期刊介绍:
Cell Host & Microbe is a scientific journal that was launched in March 2007. The journal aims to provide a platform for scientists to exchange ideas and concepts related to the study of microbes and their interaction with host organisms at a molecular, cellular, and immune level. It publishes novel findings on a wide range of microorganisms including bacteria, fungi, parasites, and viruses. The journal focuses on the interface between the microbe and its host, whether the host is a vertebrate, invertebrate, or plant, and whether the microbe is pathogenic, non-pathogenic, or commensal. The integrated study of microbes and their interactions with each other, their host, and the cellular environment they inhabit is a unifying theme of the journal. The published work in Cell Host & Microbe is expected to be of exceptional significance within its field and also of interest to researchers in other areas. In addition to primary research articles, the journal features expert analysis, commentary, and reviews on current topics of interest in the field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


