用于疫苗和免疫疗法临床前评估的佐剂数据库
IF 7.2
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
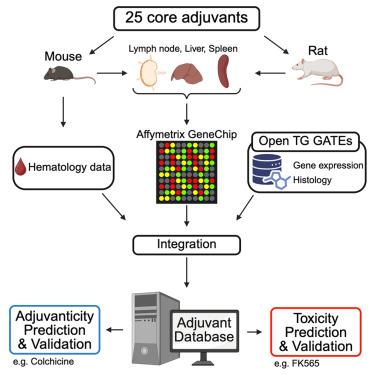
佐剂是免疫刺激剂,用于增强疫苗对传染病的效力。然而,目前评估其有效性和安全性的方法有限,阻碍了大规模筛查。为了解决这个问题,我们开发了一个包含转录组数据的原型佐剂数据库(ADB),使用与广泛使用的Open TG-GATEs (OTG)毒物基因组学数据库相同的协议生成,涵盖了跨越多个物种、器官、时间点和剂量的25种佐剂。这实现了ADB和OTG的跨数据库集成。转录组学模式成功地区分了每种佐剂,而不考虑器官或物种。使用这两个数据库,我们建立了机器学习模型来预测佐剂性和肝毒性。值得注意的是,我们通过数据驱动分析确定了秋水仙碱的佐剂活性和FK565的肝毒性。总体而言,ADB与OTG的结合为基于转录组学、数据驱动的佐剂候选筛选提供了一个框架。本文章由计算机程序翻译,如有差异,请以英文原文为准。

An adjuvant database for preclinical evaluation of vaccines and immunotherapeutics
Adjuvants are immunostimulators used to enhance vaccine efficacy against infectious diseases. However, current methods for evaluating their efficacy and safety are limited, hindering large-scale screening. To address this, we developed a prototype Adjuvant Database (ADB) containing transcriptome data, generated using the same protocols as the widely used Open TG-GATEs (OTG) toxicogenomics database, covering 25 adjuvants across multiple species, organs, time points, and doses. This enabled cross-database integration of ADB and OTG. Transcriptomic patterns successfully distinguished each adjuvant regardless of organs or species. Using both databases, we built machine learning models to predict adjuvanticity and hepatotoxicity. Notably, we identified colchicine’s adjuvant activity and FK565’s liver toxicity through data-driven analysis. Overall, ADB combined with OTG offers a framework for transcriptomics-based, data-driven screening of adjuvant candidates.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Chemical Biology
Biochemistry, Genetics and Molecular Biology-Molecular Medicine
CiteScore
14.70
自引率
2.30%
发文量
143
期刊介绍:
Cell Chemical Biology, a Cell Press journal established in 1994 as Chemistry & Biology, focuses on publishing crucial advances in chemical biology research with broad appeal to our diverse community, spanning basic scientists to clinicians. Pioneering investigations at the chemistry-biology interface, the journal fosters collaboration between these disciplines. We encourage submissions providing significant conceptual advancements of broad interest across chemical, biological, clinical, and related fields. Particularly sought are articles utilizing chemical tools to perturb, visualize, and measure biological systems, offering unique insights into molecular mechanisms, disease biology, and therapeutics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


