异常剪接导致ALS/FTD中C9orf72重复扩增
IF 20
1区 医学
Q1 NEUROSCIENCES
引用次数: 0
摘要
C9orf72 (C9)基因第一个注释内含子内的核苷酸重复扩增(NRE) (GGGGCC)n是肌萎缩性侧索硬化症(ALS)和额颞叶痴呆(FTD)的常见原因。虽然先前的研究表明C9 NRE产生几种有毒的二肽重复(DPR)蛋白,但内含子RNA片段进入细胞质翻译机制的机制尚不清楚。通过选择性地从患者来源的成纤维细胞和神经元中捕获和测序含有NRE的rna (NRE-capture-seq),我们发现,与之前的模型相比,由于使用了各种下游替代5 '剪接位点,C9 NRE作为延长的外显子1的一部分被保留。这些异常剪接异构体在C9-ALS/FTD大脑中积累,它们的产生是由富含丝氨酸/精氨酸的剪接因子1 (SRSF1)促进的。针对SRSF1或异常C9剪接异构体的反义寡核苷酸降低了DPR的水平。总之,我们的研究结果揭示了异常剪接在含nre rna的生物发生中的关键作用,并展示了针对这些致病转录物的潜在治疗策略。本文章由计算机程序翻译,如有差异,请以英文原文为准。
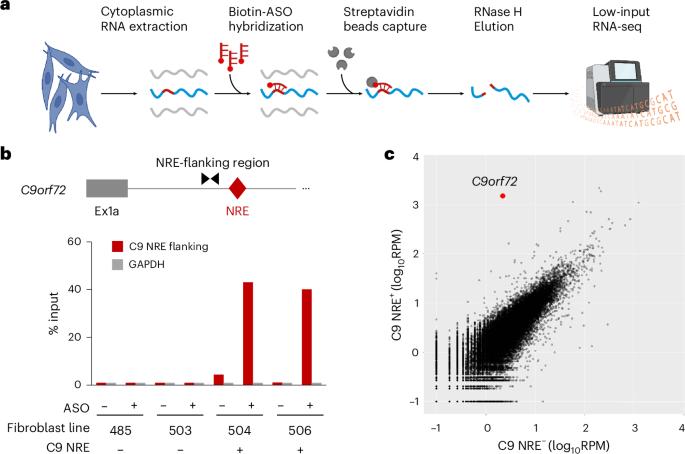
Aberrant splicing exonizes C9orf72 repeat expansion in ALS/FTD
A nucleotide repeat expansion (NRE) (GGGGCC)n within the first annotated intron of the C9orf72 (C9) gene is a common cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). While previous studies have shown that C9 NRE produces several toxic dipeptide repeat (DPR) proteins, the mechanism by which an intronic RNA segment can access the cytoplasmic translation machinery remains unclear. By selectively capturing and sequencing NRE-containing RNAs (NRE-capture-seq) from patient-derived fibroblasts and neurons, we found that, in contrast to previous models, C9 NRE is retained as part of an extended exon 1 due to the usage of various downstream alternative 5′ splice sites. These aberrant splice isoforms accumulate in C9-ALS/FTD brains, and their production is promoted by serine/arginine-rich splicing factor 1 (SRSF1). Antisense oligonucleotides targeting either SRSF1 or the aberrant C9 splice isoforms reduced the levels of DPR. Together, our findings revealed a crucial role of aberrant splicing in the biogenesis of NRE-containing RNAs and demonstrated potential therapeutic strategies to target these pathogenic transcripts. By selectively isolating and sequencing the rare RNA transcripts containing C9orf72 repeat expansion from ALS–FTD neurons, the authors uncover an alternative splicing mechanism that explains the retention of this intron segment in a translated mRNA.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature neuroscience
医学-神经科学
CiteScore
38.60
自引率
1.20%
发文量
212
审稿时长
1 months
期刊介绍:
Nature Neuroscience, a multidisciplinary journal, publishes papers of the utmost quality and significance across all realms of neuroscience. The editors welcome contributions spanning molecular, cellular, systems, and cognitive neuroscience, along with psychophysics, computational modeling, and nervous system disorders. While no area is off-limits, studies offering fundamental insights into nervous system function receive priority.
The journal offers high visibility to both readers and authors, fostering interdisciplinary communication and accessibility to a broad audience. It maintains high standards of copy editing and production, rigorous peer review, rapid publication, and operates independently from academic societies and other vested interests.
In addition to primary research, Nature Neuroscience features news and views, reviews, editorials, commentaries, perspectives, book reviews, and correspondence, aiming to serve as the voice of the global neuroscience community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


