水平获得的脂肪酸去饱和酶的进化使弹线虫能够生物合成ω-3多不饱和脂肪酸。
IF 3.7
2区 农林科学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
海洋生态系统富含必需膳食脂肪酸,特别是ω-3长链多不饱和脂肪酸(PUFAs),其中包括二十碳五烯酸(EPA)。包括微藻在内的各种海洋微生物通过甲基和前端去饱和酶的作用产生ω-3长链PUFAs。然而,由于缺乏甲基和/或前端去饱和酶,大多数陆生生物不能产生ω-3长链pufa。一个值得注意的例外是在弹尾虫(弹尾虫)中重新合成EPA,这是一种非昆虫的土壤六足动物,是掠食性动物的营养来源;然而,线虫的EPA生物合成酶仍不为人所知。在这里,我们提供了线虫生物合成EPA所需的关键去饱和酶的第一个证据。质谱分析表明,EPA是通过花生四烯酸生物合成和ω-3脱饱和合成的,ω-3脱饱和分别需要前端和甲基端脱饱和酶。编码前端去饱和酶的多候选弹线虫转录本的rna测序然而,没有检索到甲基端去饱和酶的明确同源序列。利用酵母表达系统对分离酶的活性进行验证,发现弹线虫前端去饱和酶催化花生四烯酸的生物合成。此外,弹线虫前端去饱和酶序列的一个亚群催化ω-3去饱和步骤,促进花生四烯酸向EPA的生物转化。系统发育分析进一步表明,弹线虫前端去饱和酶序列与产生ω-3长链PUFAs的海洋微生物的去饱和酶序列密切相关。鉴于弹珠虫起源于海洋甲壳类动物祖先,我们认为从海洋微生物中水平获取前端去饱和酶基因,然后进行复制和新功能化,使弹珠虫成为EPA生产者。本文章由计算机程序翻译,如有差异,请以英文原文为准。
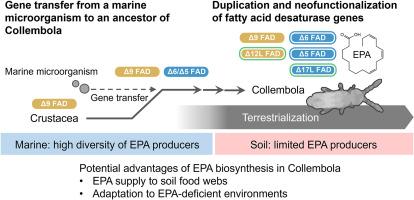
Evolution of a horizontally acquired fatty acid desaturase enables the biosynthesis of ω-3 polyunsaturated fatty acids in Collembola
Marine ecosystems are rich in essential dietary fatty acids, particularly ω-3 long-chain polyunsaturated fatty acids (PUFAs) including eicosapentaenoic acid (EPA). Various marine microorganisms including microalgae produce ω-3 long-chain PUFAs through the action of methyl- and front-end desaturases. However, most terrestrial organisms cannot produce ω-3 long-chain PUFAs due to the absence of methyl- and/or front-end desaturases. One notable exception is de novo EPA biosynthesis in springtails (Collembola), the non-insect soil hexapod lineage serving as a nutrient source for predatory animals; however, Collembola EPA biosynthesis enzymes remain unknown. Here, we provide the first evidence of the key desaturases required for EPA biosynthesis in Collembola. Mass spectrometric analysis suggested that EPA is synthesized through arachidonic acid biosynthesis and following ω-3 desaturation, which requires front- and methyl-end desaturases, respectively. RNA-sequencing of Collembola transcripts isolated multiple candidate genes encoding front-end desaturases; however, no clear orthologous sequence of a methyl-end desaturase was retrieved. Verification of the activity of the isolated enzymes using a yeast expression system revealed that Collembola front-end desaturases catalyzed arachidonic acid biosynthesis. Furthermore, a subgroup of Collembola front-end desaturase sequences catalyzed the ω-3 desaturation step, facilitating the bioconversion of arachidonic acid to EPA. Phylogenetic analysis further revealed that the Collembola front-end desaturase sequences clustered closely to those of marine microorganisms producing ω-3 long-chain PUFAs. Given that Collembola is derived from a marine Crustacea ancestor, we propose that the horizontal acquisition of a front-end desaturase gene from a marine microbe, followed by duplication and neofunctionalization, empowered Collembola to become an EPA producer.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
7.40
自引率
5.30%
发文量
105
审稿时长
40 days
期刊介绍:
This international journal publishes original contributions and mini-reviews in the fields of insect biochemistry and insect molecular biology. Main areas of interest are neurochemistry, hormone and pheromone biochemistry, enzymes and metabolism, hormone action and gene regulation, gene characterization and structure, pharmacology, immunology and cell and tissue culture. Papers on the biochemistry and molecular biology of other groups of arthropods are published if of general interest to the readership. Technique papers will be considered for publication if they significantly advance the field of insect biochemistry and molecular biology in the opinion of the Editors and Editorial Board.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


