SWIFT-seq能够对多发性骨髓瘤及其前体的循环肿瘤细胞进行全面的单细胞转录组学分析。
IF 28.5
1区 医学
Q1 ONCOLOGY
引用次数: 0
摘要
多发性骨髓瘤是一种骨髓浆细胞恶性肿瘤,有前体病变。脑脊髓瘤活检很少进行,由于技术限制,可能产生不确定的结果。循环肿瘤细胞(ctc)谱分析可以实现无创常规临床评估,但仍然具有挑战性。在这里,为了解决这个问题,我们描述了一个单细胞测序工作流程来询问少量肿瘤细胞(SWIFT-seq),并对来自101例患者和健康供体的配对BM和ctc使用单细胞RNA测序和B细胞受体测序。我们建立了基于序列的CTC枚举策略,并开发了CTC分类器来推断细胞遗传学异常。此外,我们利用表达谱来测量CTCs中的肿瘤增殖指数,并证明在CTCs中可以捕获克隆动态。最后,我们提出了一个循环动力学模型,其中肿瘤负荷、增殖、细胞遗传学和循环能力特征影响CTC负荷。总之,SWIFT-seq可能会促进基于血液的骨髓瘤诊断、监测和预后,并揭示肿瘤传播的生物学机制。本文章由计算机程序翻译,如有差异,请以英文原文为准。
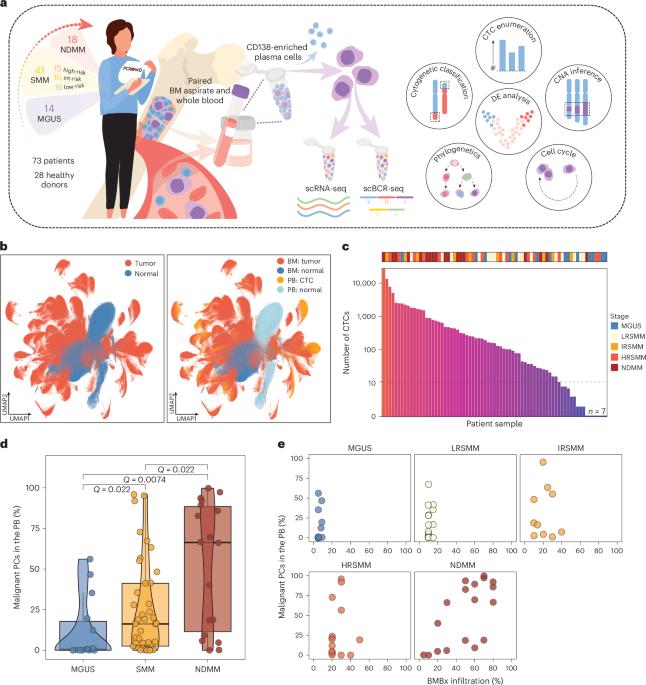
SWIFT-seq enables comprehensive single-cell transcriptomic profiling of circulating tumor cells in multiple myeloma and its precursors
Multiple myeloma is a bone marrow (BM) plasma cell malignancy preceded by precursor conditions. BM biopsies are conducted infrequently and can yield inconclusive results due to technical limitations. Profiling circulating tumor cells (CTCs) may enable noninvasive routine clinical assessments but remains challenging. Here, to address this, we describe a single-cell sequencing workflow to interrogate few tumor cells (SWIFT-seq), and employ single-cell RNA sequencing and B cell receptor sequencing on paired BM and CTCs from 101 patients and healthy donors. We establish a sequencing-based CTC enumeration strategy and develop a CTC classifier to infer cytogenetic abnormalities. Additionally, we leverage expression profiling to measure tumor proliferative index in CTCs, and demonstrate that clonal dynamics can be captured in CTCs. Last, we propose a circulatory dynamics model whereby tumor burden, proliferation, cytogenetics and a circulatory capacity signature influence CTC burden. Overall, SWIFT-seq may advance blood-based myeloma diagnostics, surveillance and prognostication, and reveal biological mechanisms of tumor dissemination. Lightbody et al. present SWIFT-seq, a single-cell RNA sequencing approach to profile circulating tumor cells from patients with multiple myeloma and its precursor conditions and leverage it to derive clinical and biological insights into the disease.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature cancer
Medicine-Oncology
CiteScore
31.10
自引率
1.80%
发文量
129
期刊介绍:
Cancer is a devastating disease responsible for millions of deaths worldwide. However, many of these deaths could be prevented with improved prevention and treatment strategies. To achieve this, it is crucial to focus on accurate diagnosis, effective treatment methods, and understanding the socioeconomic factors that influence cancer rates.
Nature Cancer aims to serve as a unique platform for sharing the latest advancements in cancer research across various scientific fields, encompassing life sciences, physical sciences, applied sciences, and social sciences. The journal is particularly interested in fundamental research that enhances our understanding of tumor development and progression, as well as research that translates this knowledge into clinical applications through innovative diagnostic and therapeutic approaches. Additionally, Nature Cancer welcomes clinical studies that inform cancer diagnosis, treatment, and prevention, along with contributions exploring the societal impact of cancer on a global scale.
In addition to publishing original research, Nature Cancer will feature Comments, Reviews, News & Views, Features, and Correspondence that hold significant value for the diverse field of cancer research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


