人胎儿组织RNA-m5C调控图谱揭示了Nsun2/Jarid2/Alyref轴的活性
IF 13
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
胞嘧啶甲基化(m5C)是人类胎儿发育中必不可少的关键RNA修饰,但其组织特异性分布和调控机制仍未明确。目的构建m5C在人胎儿组织中的组织特异性图谱,分析RNA甲基化与表观遗传调控之间的表观遗传串扰。具体来说,我们关注的是nsun2介导的m5C修饰如何影响胎儿发育过程中染色质动力学和组蛋白修饰模式。方法对7种不同的人类胎儿组织进行亚硫酸氢盐RNA测序(m5C-Seq)和转录组分析(RNA- seq)。为了探索m5C的功能作用,我们采用了Nsun2条件敲除和Nsun5敲除小鼠模型。我们进一步整合了多种实验方法,包括点印迹法、免疫荧光显微镜、Co-IP、CUT&;Tag和ATAC-Seq,以研究m5C修饰、组蛋白修饰和染色质可及性之间的功能联系。我们建立了人类胎儿组织中m5C修饰的第一个综合图谱,揭示了不同的组织特异性甲基化模式。我们进一步证明,m5c修饰的转录物维持基因表达稳态,并通过选择性剪接调节关键的发育转录程序。在机制上,我们发现Nsun2招募Jarid2/Ezh2复合体,并通过Alyref的m5c依赖性识别调节其活性。该通路协调组蛋白修饰和染色质可及性,支持胎儿正常发育。结论本研究提供了人类胎儿发育过程中高分辨率、全面的组织特异性m5C图谱,为发育表观遗传学研究提供了基础资源。随后,我们发现了一个连接Nsun2、Jarid2/Ezh2复合物、m5C修饰模式和Alyref功能的新调控轴。该途径整合了RNA甲基化、组蛋白修饰和染色质可及性,以精确地协调胎儿发育,促进了我们对表观遗传调控的理解,并为发育障碍的潜在治疗靶点提供了新的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
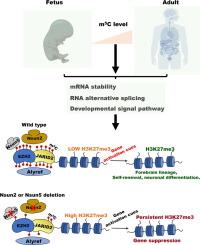
RNA-m5C regulatory atlas of human fetal tissues uncover the activities of Nsun2/Jarid2/Alyref axis
Introduction
Cytosine methylation (m5C) is a pivotal RNA modification essential for human fetal development, yet its tissue-specific distribution and regulatory mechanisms remain largely undefined.Objectives
This study aimed to construct a tissue-specific atlas of m5C distribution across human fetal tissues and to dissect the epigenetic crosstalk between RNA methylation and epigenetic regulation. Specifically, we focused on how Nsun2-mediated m5C modifications influence chromatin dynamics and histone modification patterns during fetal development.Methods
We performed RNA bisulfite sequencing (m5C-Seq) and transcriptome profiling (RNA-Seq) across seven distinct human fetal tissues. To explore the functional roles of m5C, we employed Nsun2 conditional knockout and Nsun5 knockout mouse models. We further integrated multiple experimental approaches, including dot blot assays, immunofluorescence microscopy, Co-IP, CUT&Tag and ATAC-Seq, to examine the functional connections between m5C modification, histone modifications and chromatin accessibility.Results
We established the first comprehensive atlas of m5C modifications across human fetal tissues, revealing distinct tissue-specific methylation patterns. We further demonstrated that m5C-modified transcripts maintain gene expression homeostasis and regulate critical developmental transcriptional programs through alternative splicing. Mechanistically, we found that Nsun2 recruits the Jarid2/Ezh2 complex and regulates its activity via m5C-dependent recognition by Alyref. This pathway coordinates histone modifications and chromatin accessibility, supporting proper fetal development.Conclusion
Our study presents a high-resolution, comprehensive tissue-specific m5C landscape during human fetal development, serving as a foundational resource for developmental epigenetics research. Subsequently, we identified a novel regulatory axis linking Nsun2, the Jarid2/Ezh2 complex, m5C modification patterns and Alyref functionality. This pathway integrates RNA methylation, histone modification and chromatin accessibility to precisely orchestrate fetal development, advancing our understanding of epigenetic regulation and offering new insights into potential therapeutic targets for developmental disorders.求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


