内源性小脑底物的检测和富集方法
IF 7.2
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
c端环亚胺是蛋白质上的翻译后修饰(PTMs),被E3连接酶底物适配器小脑(CRBN)识别和去除。尽管通过基于质谱的蛋白质组学观察到蛋白质组中的这些修饰,但一种正交和可推广的方法来可视化c端环亚胺将提高内源性CRBN底物表征的检测、灵敏度和通量。在这里,我们开发了一种抗体样试剂,称为“cerebody”,用于可视化和富集c端环亚胺修饰的蛋白质。我们描述了CRBN衍生物的工程,以产生小脑体,并使用它来识别CRBN底物,通过western blot和富集全细胞和组织裂解物。通过小脑富集鉴定的CRBN底物被定位、验证,并进一步表征其依赖于c端环亚胺修饰。这些方法将加速内源性CRBN底物的表征及其调控。本文章由计算机程序翻译,如有差异,请以英文原文为准。
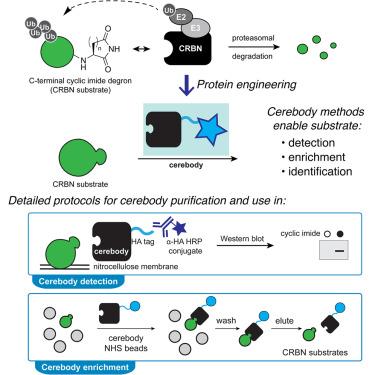
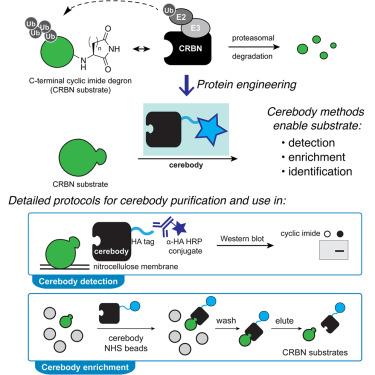
A method for the detection and enrichment of endogenous cereblon substrates
C-terminal cyclic imides are posttranslational modifications (PTMs) on proteins that are recognized and removed by the E3 ligase substrate adapter cereblon (CRBN). Despite the observation of these modifications across the proteome by mass spectrometry-based proteomics, an orthogonal and generalizable method to visualize the C-terminal cyclic imide would enhance detection, sensitivity, and throughput of endogenous CRBN substrate characterization. Here, we develop an antibody-like reagent, termed “cerebody,” for visualizing and enriching C-terminal cyclic imide-modified proteins. We describe the engineering of CRBN derivatives to produce cerebody and use it to identify CRBN substrates by western blot and enrichment from whole-cell and tissue lysates. CRBN substrates identified by cerebody enrichment are mapped, validated, and further characterized for dependence on the C-terminal cyclic imide modification. These methods will accelerate the characterization of endogenous CRBN substrates and their regulation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell Chemical Biology
Biochemistry, Genetics and Molecular Biology-Molecular Medicine
CiteScore
14.70
自引率
2.30%
发文量
143
期刊介绍:
Cell Chemical Biology, a Cell Press journal established in 1994 as Chemistry & Biology, focuses on publishing crucial advances in chemical biology research with broad appeal to our diverse community, spanning basic scientists to clinicians. Pioneering investigations at the chemistry-biology interface, the journal fosters collaboration between these disciplines. We encourage submissions providing significant conceptual advancements of broad interest across chemical, biological, clinical, and related fields. Particularly sought are articles utilizing chemical tools to perturb, visualize, and measure biological systems, offering unique insights into molecular mechanisms, disease biology, and therapeutics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


