核心剪接结构和早期剪接体识别决定了微外显子对SRRM3/4的敏感性
IF 10.1
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
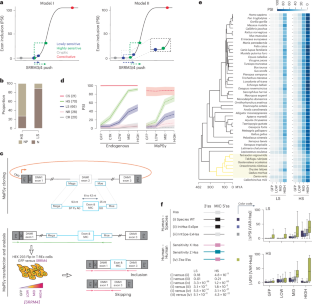
微外显子对于神经元和胰腺内分泌细胞的正常运作至关重要,它们的包含取决于剪接因子SRRM3和SRRM4 (SRRM3/4)。然而,在胰腺细胞中,这些调节因子的低表达限制了所有神经元微外显子中仅包含最敏感的亚群。尽管多种顺式作用元件可参与微外显子调控,但它们如何决定这种差异剂量反应以及相应的对SRRM3/4的高或低敏感性尚不清楚。在这里,我们使用大量平行剪接试验探测28,535个变体,以显示对SRRM4的敏感性在脊椎动物中是保守的。我们的数据支持一个调控模型,即微外显子的高或低灵敏度在很大程度上取决于核心剪接结构和长度约束之间的相互作用。在SRRM3/4缺失的情况下,不同的剪接体活性进一步支持了这一结论,并通过数学模型假设这两种类型的微外显子仅在招募早期剪接体成分的效率上有所不同。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Core splicing architecture and early spliceosomal recognition determine microexon sensitivity to SRRM3/4
Microexons are essential for the proper operation of neurons and pancreatic endocrine cells, in which their inclusion depends on the splicing factors SRRM3 and SRRM4 (SRRM3/4). However, in pancreatic cells, lower expression of these regulators limits inclusion to only the most sensitive subset among all neuronal microexons. Although various cis-acting elements can contribute to microexon regulation, how they determine this differential dose response and the corresponding high or low sensitivity to SRRM3/4 remains unknown. Here we use massively parallel splicing assays probing 28,535 variants to show that sensitivity to SRRM4 is conserved across vertebrates. Our data support a regulatory model whereby high or low microexon sensitivity is largely determined by the interplay between core splicing architecture and length constraints. This conclusion is further supported by distinct spliceosome activities in the absence of SRRM3/4 and by a mathematical model that assumes that the two types of microexons differ only in their efficiency to recruit early spliceosomal components. Using massively parallel splicing assays and mathematical modeling, Bonnal et al. uncover that conserved splice site strength and exon length encode microexon sensitivity to SRRM3 and SRRM4. This work revises the switch-like splicing regulation model into a dose-responsive continuum.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


