通过人工智能辅助管道鉴定肠道微生物胆汁酸代谢酶
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
胆汁酸(BAs)的修饰是其在宿主生理和病理中的作用的基础。确定它们的合成酶对于揭示BAs的多样性和开发有针对性的干预措施至关重要,但这仍然是一个重大挑战。为了解决这个问题,我们开发了一个人工智能(AI)辅助的工作流程,胆汁酸酶播音员单元工具(BEAUT),该工具预测了超过60万种候选BA代谢酶,并将其编译到人类通用微生物BA代谢酶(HGBME)数据库(https://beaut.bjmu.edu.cn)中。我们鉴定了一系列未被鉴定的BA酶,包括单酸酰化BA水解酶(MABH)和3-乙酰dca合成酶(ADS)。值得注意的是,ADS可以产生未报道的骨架BA, 3-acetoDCA,具有碳-碳键延伸。在确定其细菌来源和催化机制后,我们发现3-acetoDCA广泛分布于人群中,并调节肠道内微生物的相互作用。总之,我们的工作从酶的角度为微生物BAs和宿主之间的关系提供了另一种见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
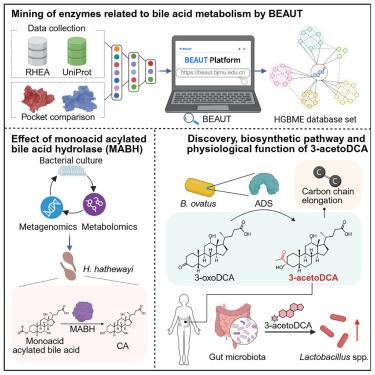
Identification of gut microbial bile acid metabolic enzymes via an AI-assisted pipeline
The modifications of bile acids (BAs) are fundamental to their role in host physiology and pathology. Identifying their synthetases is crucial for uncovering the diversity of BAs and developing targeted interventions, yet it remains a significant challenge. To address this hurdle, we developed an artificial intelligence (AI)-assisted workflow, bile acid enzyme announcer unit tool (BEAUT), which predicted over 600,000 candidate BA metabolic enzymes that we compiled into the human generalized microbial BA metabolic enzyme (HGBME) database (https://beaut.bjmu.edu.cn). We identified a series of uncharacterized BA enzymes, including monoacid acylated BA hydrolase (MABH) and 3-acetoDCA synthetase (ADS). Notably, ADS can produce an unreported skeleton BA, 3-acetoDCA, with a carbon-carbon bond extension. After determining its bacterial source and catalytic mechanism, we found that 3-acetoDCA is widely distributed among populations and regulates the microbial interactions in the gut. In conclusion, our work offers alternative insights into the relationship between microbial BAs and the host from an enzymatic perspective.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


