单倍型解决,无间隙的基因组组装提供了对亚洲和欧洲梨之间差异的见解
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
梨(Pyrus spp.)是具有广泛遗传多样性的自交不亲和作物。高杂合性和技术限制导致参考基因组之间存在缺口。我们的研究展示了具有代表性的亚洲梨“砀山苏里”(Pyrus bretschneideri)和欧洲梨“Max Red Bartlett”(Pyrus communis)的端粒到端粒、单倍型解析基因组。单倍型特异性基因与双等位基因在转座因子含量、甲基化模式和表达水平上存在显著差异。等位基因特异性表达分析表明,显性效应对梨果实品质的形成起着至关重要的作用。对362份材料的种群分析表明,种间渐渗增加了梨的多样性。我们构建了一个基于图的基因组,并确定了与农艺性状相关的结构变异。在亚洲和欧洲梨中发现了一个286 bp的启动子区域插入,以及PyACS1的差异表达,它们具有不同的果实软化特性。进一步的实验证实了PyACS1在水果软化中的作用。总的来说,这项研究提供了遗传变异的见解,并将促进梨的改良。本文章由计算机程序翻译,如有差异,请以英文原文为准。
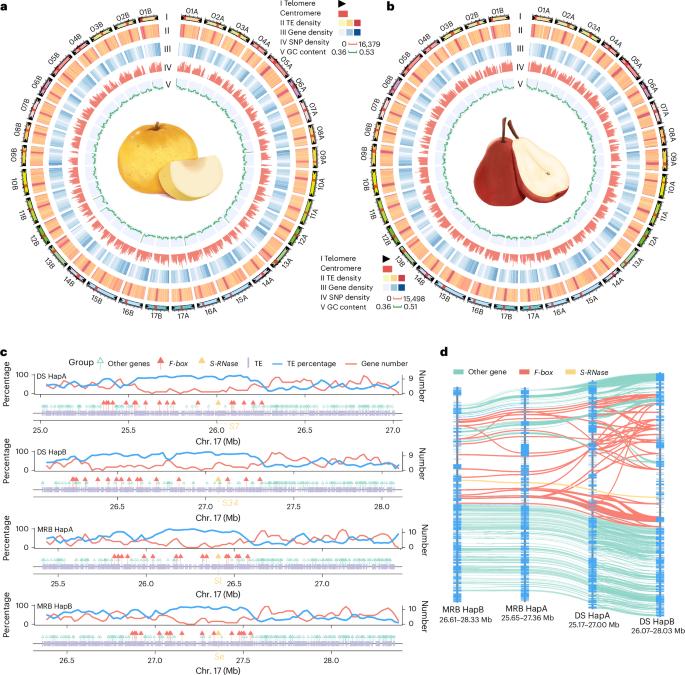
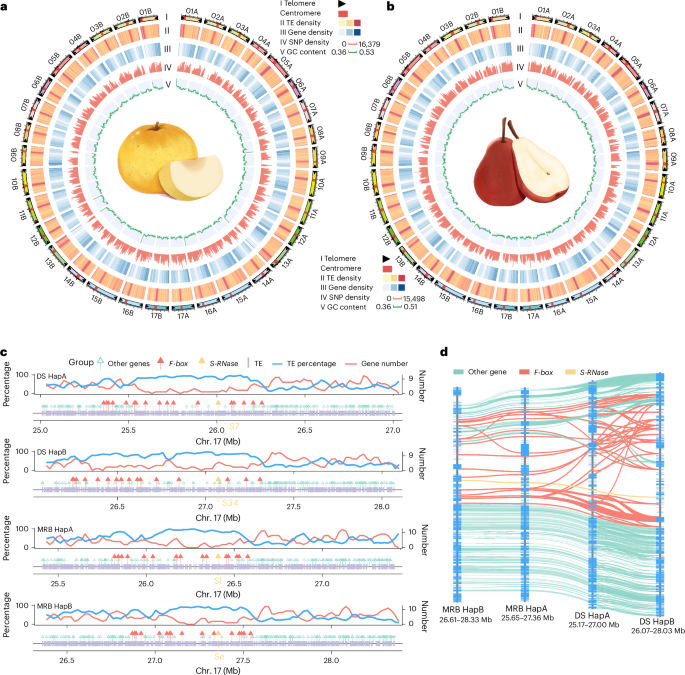
Haplotype-resolved, gap-free genome assemblies provide insights into the divergence between Asian and European pears
Pears (Pyrus spp.) are self-incompatible crops with broad genetic diversity. High heterozygosity and technical limitations result in gaps within reference genomes. Our study presents the telomere-to-telomere, haplotype-resolved genomes of a representative Asian pear ‘Dangshansuli’ (Pyrus bretschneideri) and a European pear ‘Max Red Bartlett’ (Pyrus communis). Haplotype-specific genes exhibited notable differences from biallelic genes regarding transposable content, methylation patterns and expression levels. Allele-specific expression analysis suggested that the dominance effect is vital in the formation of fruit quality of pears. Population analysis of 362 accessions revealed that interspecific introgression increased pear diversity. We constructed a graph-based genome and identified structural variations associated with agronomic traits. A 286-bp insertion in the promoter region, along with differential expression of PyACS1, was identified between Asian and European pears, which exhibit distinct fruit-softening characteristics. Further experiments demonstrated the role of PyACS1 in fruit softening. Overall, this study provided insights into genetic variation and will facilitate pear improvement. Haplotype-resolved, gap-free genome assemblies for a representative Asian pear (Pyrus bretschneideri, ‘Dangshansuli’) and a European pear (Pyrus communis, ‘Max Red Bartlett’) provide insights into genome evolution and interspecies variation in Pyrus species.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


