高光谱成像技术在豌豆根腐病早期检测中的应用
IF 3.3
3区 农林科学
Q2 PLANT SCIENCES
引用次数: 0
摘要
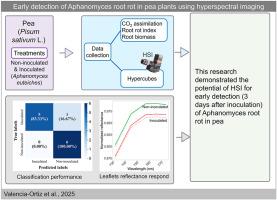
植物与病原体相互作用产生的植物叶片和根系反射可以提供有关疾病状态的信息,使它们对早期疾病检测有用。本研究在控制条件下,利用高光谱成像(HSI)技术评价了豌豆(Pisum sativum L.)植株接种引起根腐病(Aphanomyces root rot, ARR)的病原菌euteiches Drechs后的早期反应。在水培条件下培养具有数量性状位点Ae - Ps7.6的2个ARR部分抗性品系(NIL5-7.6b和NIL8-7.6b)和相应的对照(NIL5-0b和NIL8-0b,不含QTL),采用裂地设计,采用未接种和接种(1 × 105个zoo孢子ml - 1)两种处理,共6个重复。HSI数据采集于接种后3天最年轻的小叶(DAI)。在8 DAI时,收集根的HSI数据。对小叶和根的HSI超立方体进行处理,去除背景,提取每个样本跨波长的平均值。利用小叶高光谱特征计算归一化差分光谱指数。然后,采用交叉验证递归特征消除和随机森林分类器选择重要特征并进行推理分析。对于根数据,使用了类似的方法,但是,选择的重要特征在随机森林和梯度增强分类器中使用。小叶结果表明,745 nm红边波长是3 DAI处理可分离性的基本特征。同时,在随机森林和梯度增强的情况下,根分析的分类准确率分别达到83%和92%。这项研究为HSI检测ARR的潜力提供了有价值的见解,特别是在植物疾病症状前的早期阶段。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Early detection of Aphanomyces root rot in pea plants using hyperspectral imaging
Plant leaf and root reflectance resulting from plant–pathogen interaction can be informative about disease status, making them useful for early disease detection. In controlled conditions, this research utilized a hyperspectral imaging (HSI) system to evaluate the early response of pea plants (Pisum sativum L.) inoculated with Aphanomyces euteiches Drechs, the causal agent of Aphanomyces root rot (ARR). Two ARR partially resistant lines (NIL5-7.6b and NIL8-7.6b) with the quantitative trait locus (QTL) Ae - Ps7.6 and corresponding controls (NIL5-0b and NIL8-0b, without QTL) were grown in hydroponic conditions and organized in a split-plot design using two treatments, non-inoculated and inoculated (1 × 105 zoospores ml−1) with six replications. The HSI data were collected from the youngest leaflets 3 days after inoculation (DAI). At 8 DAI, HSI data from roots were collected. The HSI hypercubes of leaflets and roots were processed to remove the background and extract the mean value of each sample across wavelengths. Leaflet hyperspectral signatures were used to calculate normalized difference spectral indices. Then, a recursive feature elimination with cross-validation and a random forest classifier was used to select important features and test them with inferential analysis. For root data, a similar approach was used, however, the selected important features were used in random forest and gradient boosting classifiers. The leaflet results showed the red-edge wavelength of 745 nm was an essential feature for treatment separability at 3 DAI. Meanwhile, root analysis displayed a high classification accuracy of 83% and 92% with random forest and gradient boosting, respectively. This research offers valuable insights into the potential of HSI for ARR detection, particularly in the early pre-symptomatic stages of the plant disease.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
4.30
自引率
7.40%
发文量
130
审稿时长
38 days
期刊介绍:
Physiological and Molecular Plant Pathology provides an International forum for original research papers, reviews, and commentaries on all aspects of the molecular biology, biochemistry, physiology, histology and cytology, genetics and evolution of plant-microbe interactions.
Papers on all kinds of infective pathogen, including viruses, prokaryotes, fungi, and nematodes, as well as mutualistic organisms such as Rhizobium and mycorrhyzal fungi, are acceptable as long as they have a bearing on the interaction between pathogen and plant.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


