单核染色质可及性分析鉴定导致重度抑郁症的细胞类型和功能变异
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
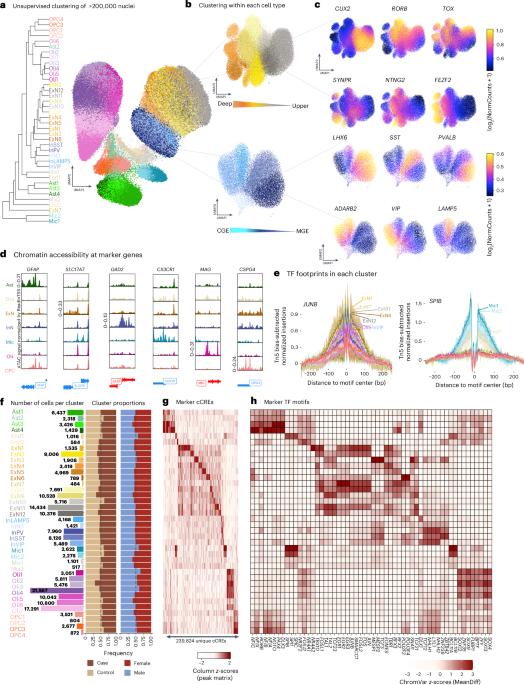
与重度抑郁症(MDD)相关的遗传变异在调控基因组中富集。在这里,我们通过将84个个体的背外侧前额叶皮层的200,000多个细胞的单细胞染色质可及性与基因表达结合起来,研究了MDD与神经正常对照相比的基因调控机制。mdd相关的染色质可及性改变在以转录因子(TF)基序可及性和NR4A2结合为特征的深层兴奋性神经元中表现突出,NR4A2是一种对应激有反应的活性依赖的TF。同样的神经元富含mdd相关的遗传变异,破坏了与可能影响突触通讯的基因相关的TF结合位点。此外,在MDD患者中,灰质小胶质细胞簇在已知调节免疫稳态的tf结合位点的可及性降低。最后,我们通过基于序列的可及性预测、供体特异性基因型和基于细胞的测定,确定了mdd风险变异的基因调控作用。这些发现揭示了基因变异可能增加MDD风险的细胞类型和调控机制。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Single-nucleus chromatin accessibility profiling identifies cell types and functional variants contributing to major depression
Genetic variants associated with major depressive disorder (MDD) are enriched in the regulatory genome. Here, we investigate gene-regulatory mechanisms underlying MDD compared to neurotypical controls by combining single-cell chromatin accessibility with gene expression in over 200,000 cells from the dorsolateral prefrontal cortex of 84 individuals. MDD-associated alterations in chromatin accessibility were prominent in deep-layer excitatory neurons characterized by transcription factor (TF) motif accessibility and binding of NR4A2, an activity-dependent TF reactive to stress. The same neurons were enriched for MDD-associated genetic variants, disrupting TF binding sites linked to genes that likely affect synaptic communication. Furthermore, a gray matter microglia cluster exhibited decreased accessibility in individuals with MDD at binding sites bound by TFs known to regulate immune homeostasis. Finally, we identified gene-regulatory effects of MDD-risk variants using sequence-based accessibility predictions, donor-specific genotypes and cell-based assays. These findings shed light on the cell types and regulatory mechanisms through which genetic variation may increase the risk of MDD. Integration of snATAC-seq and snRNA-seq data from brains of individuals with major depressive disorder identifies chromatin accessibility alterations and functional enrichment of risk variants in deep-layer excitatory neurons. Gray matter microglia in these individuals show decreased accessibility at sites bound by regulators of immune homeostasis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


