迭代重组酶技术在千碱基到百万碱基尺度上高效和精确的基因组工程
IF 42.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
基因组编辑技术在实现高等生物的精确、大规模DNA操作方面面临挑战,包括低效率、有限的编辑规模和类型,以及保留不需要的序列,如重组位点(“疤痕”)。在这里,我们提出了可编程染色体工程(PCE)和RePCE,两种可编程染色体编辑系统,可以在植物和人类细胞中进行无疤痕的千碱基到百万碱基的DNA操作。通过高通量工程,我们获得了可逆性降低10倍的Lox位点,并应用ai辅助重组酶工程方法(AiCErec)生成了重组效率为野生型3.5倍的Cre变体。结合re - pegrna介导的无疤痕策略进一步提高了编辑精度,在染色体水平上允许无疤痕插入、缺失、替换、倒位和易位。主要应用包括315 kb的水稻抗除草剂反转,无疤痕染色体融合,以及12 mb的人类疾病相关位点反转。这些进展显著拓宽了基因组编辑在分子育种、治疗开发和合成生物学方面的应用范围。本文章由计算机程序翻译,如有差异,请以英文原文为准。
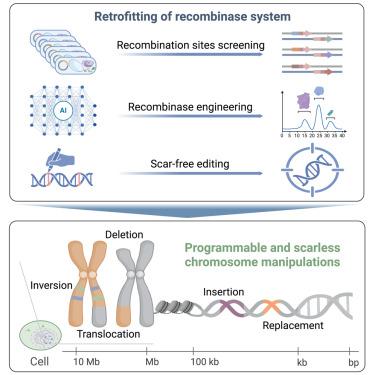
Iterative recombinase technologies for efficient and precise genome engineering across kilobase to megabase scales
Genome editing technologies face challenges in achieving precise, large-scale DNA manipulations in higher organisms, including inefficiency, limited editing scales and types, and the retention of undesired sequences such as recombination sites (“scars”). Here, we present programmable chromosome engineering (PCE) and RePCE, two programmable chromosome editing systems enabling scarless kilobase-to-megabase DNA manipulations in plants and human cells. Through high-throughput engineering, we obtained Lox sites with a 10-fold reduced reversibility and applied an AI-assisted recombinase engineering method (AiCErec) to generate Cre variants with 3.5 times the recombination efficiency of the wild type. Incorporation of a Re-pegRNA-mediated scar-free strategy further enhanced editing precision, allowing scarless insertions, deletions, replacements, inversions, and translocations at the chromosomal level. Key applications include a 315-kb inversion in rice conferring herbicide resistance, scarless chromosome fusions, and a 12-Mb inversion at human disease-related sites. These advances significantly broaden the scope of genome editing applications in molecular breeding, therapeutic development, and synthetic biology.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


