鸡胴体冲洗液中弯曲杆菌的相对离心力法计数和检测方法的验证。
IF 1.9
4区 生物学
Q4 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
弯曲杆菌病是新西兰最常见的食源性疾病,家禽是主要的感染源。新西兰根据国家微生物数据库(NMD)计划监测家禽胴体冲洗液中存在的弯曲杆菌。为了更好地监测弯曲杆菌的控制进展,需要一种更灵敏的方法来枚举弯曲杆菌数量较少的菌株。本研究对现有的NMD方法进行了改进,增加了一个相对离心力(RCF)步骤,用于从家禽胴体冲洗液中浓缩弯曲杆菌。与15 min相比,离心30 min可显著提高弯曲杆菌的回收率(p本文章由计算机程序翻译,如有差异,请以英文原文为准。
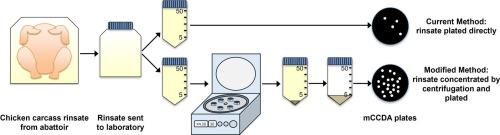
Validation of a Relative Centrifugal Force method for the enumeration and detection of Campylobacter from chicken carcass rinsates
Campylobacteriosis is the most frequently notified foodborne disease in New Zealand and poultry is the predominant infection source. New Zealand monitors Campylobacter present in poultry carcass rinsates under the National Microbiological Database (NMD) programme. To better monitor Campylobacter control improvements, a more sensitive method is required that can enumerate rinsates with lower Campylobacter numbers. This study developed a modification of the current NMD method involving adding a relative centrifugal force (RCF) step for concentrating Campylobacter from poultry carcass rinsates. Centrifugation for 30 min significantly improved Campylobacter recovery compared with 15 min (p < 0.001), but there were no differences between RCFs of 3500, 4000 and 4430 x g (p = 0.992). RCF and NMD method performances were compared in a single laboratory validation study that used different inoculation levels of twelve Campylobacter strains, including poultry isolates. Campylobacter was detected from more samples (p < 0.001) using the RCF method (93 of 126; 73.8 %) than the NMD method (65 of 126; 51.6 %). The RCF method had a seven-fold lower detection limit (28 colony forming units (CFU)/400 ml) than the NMD method (200 CFU/400 ml). The detection limit accounted for an observed 70.3 % of the inoculated CFU captured within the centrifuged pellet. Campylobacter was also detected from significantly more (p < 0.001) commercial chicken rinsate samples tested by poultry industry laboratories using the RCF method (257 of 863; 29.8 %) than the NMD method (114 of 863; 13.2 %). Taken together, results support the RCF method as a modification of the NMD method to enumerate lower numbers of Campylobacter in rinsates.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of microbiological methods
生物-生化研究方法
CiteScore
4.30
自引率
4.50%
发文量
151
审稿时长
29 days
期刊介绍:
The Journal of Microbiological Methods publishes scholarly and original articles, notes and review articles. These articles must include novel and/or state-of-the-art methods, or significant improvements to existing methods. Novel and innovative applications of current methods that are validated and useful will also be published. JMM strives for scholarship, innovation and excellence. This demands scientific rigour, the best available methods and technologies, correctly replicated experiments/tests, the inclusion of proper controls, calibrations, and the correct statistical analysis. The presentation of the data must support the interpretation of the method/approach.
All aspects of microbiology are covered, except virology. These include agricultural microbiology, applied and environmental microbiology, bioassays, bioinformatics, biotechnology, biochemical microbiology, clinical microbiology, diagnostics, food monitoring and quality control microbiology, microbial genetics and genomics, geomicrobiology, microbiome methods regardless of habitat, high through-put sequencing methods and analysis, microbial pathogenesis and host responses, metabolomics, metagenomics, metaproteomics, microbial ecology and diversity, microbial physiology, microbial ultra-structure, microscopic and imaging methods, molecular microbiology, mycology, novel mathematical microbiology and modelling, parasitology, plant-microbe interactions, protein markers/profiles, proteomics, pyrosequencing, public health microbiology, radioisotopes applied to microbiology, robotics applied to microbiological methods,rumen microbiology, microbiological methods for space missions and extreme environments, sampling methods and samplers, soil and sediment microbiology, transcriptomics, veterinary microbiology, sero-diagnostics and typing/identification.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


