ISW1和CHD1染色质重塑子抑制活酵母细胞的整体核小体动力学
IF 12.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
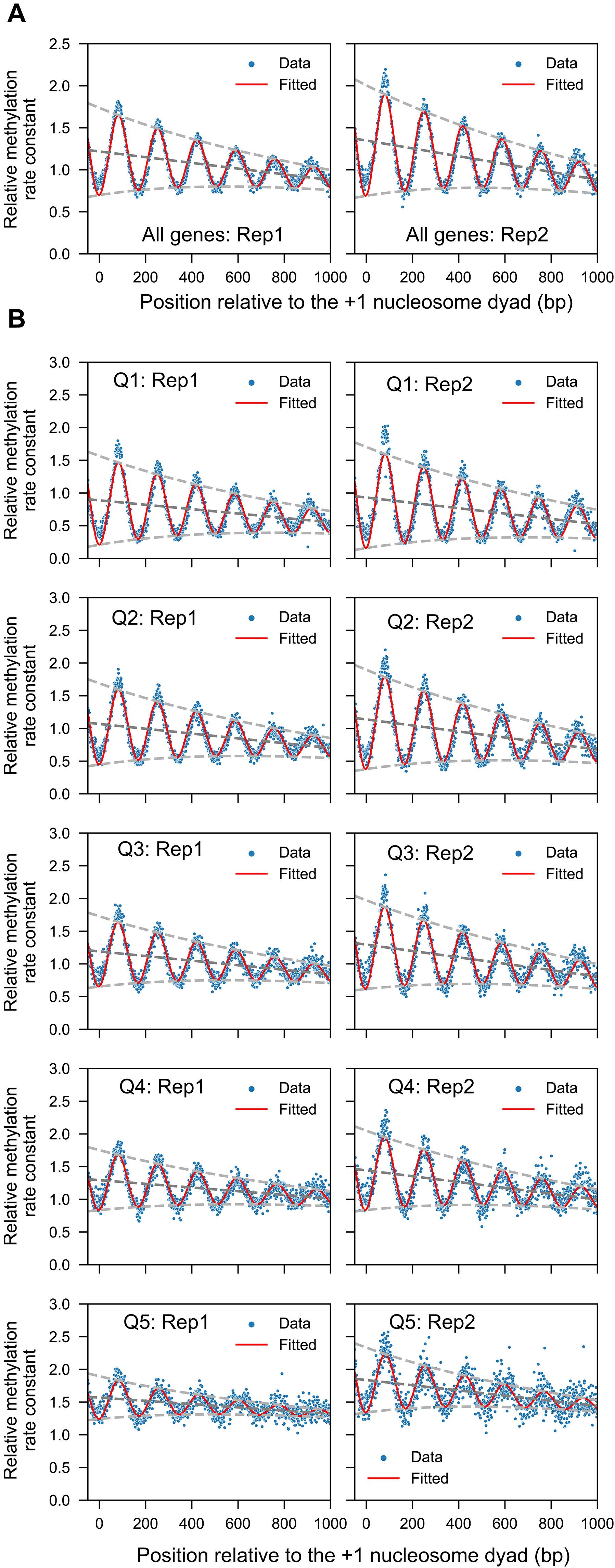
活细胞中的DNA甲基转移酶可以在全球范围内进入出芽酵母基因组,而在分离的细胞核中,DNA甲基转移酶大多无法进入。在这里,我们评估了RSC, ISW1和CHD1腺苷5 ' -三磷酸依赖的染色质重塑子在体内产生核小体动力学中的作用。我们通过将每种菌株的核甲基化率正常化到非核体线粒体DNA甲基化率来比较野生型细胞和染色质重塑突变体的DNA甲基化率。Isw1和Chd1的缺失增加了标准化甲基化率,表明这些重塑子共同抑制核小体动力学。单独消耗Isw1、Chd1或Rsc8几乎没有影响。用于拟合核小体相位数据的衰减正正波模型表明,核小体动力学以Isw1/ chd1依赖的方式随着距离启动子的距离而降低。此外,TFIIIB和TFIIIC转录因子在体内转运RNA基因时表现出不同的动力学。我们的分析提供了对体内核小体和转录因子动力学的深入了解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
The ISW1 and CHD1 chromatin remodelers suppress global nucleosome dynamics in living yeast cells
The budding yeast genome is globally accessible to DNA methyltransferases in living cells, unlike in isolated nuclei, where it is mostly inaccessible. Here, we assess the roles of the RSC, ISW1, and CHD1 adenosine 5′-triphosphate–dependent chromatin remodelers in generating nucleosome dynamics in vivo. We compare DNA methylation rates in wild-type cells and chromatin remodeler mutants by normalizing nuclear methylation rates to the nonnucleosomal mitochondrial DNA methylation rate in each strain. Depletion of both Isw1 and Chd1 increases the normalized methylation rate, suggesting that these remodelers act together to suppress nucleosome dynamics. Separate depletion of Isw1, Chd1, or Rsc8 has little effect. A decaying sine wave model used to fit nucleosome phasing data shows that nucleosome dynamics decrease with distance from the promoter in an Isw1/Chd1-dependent manner. Furthermore, the TFIIIB and TFIIIC transcription factors exhibit differential dynamics at transfer RNA genes in vivo. Our analysis provides insight into nucleosome and transcription factor dynamics in vivo.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Advances
综合性期刊-综合性期刊
CiteScore
21.40
自引率
1.50%
发文量
1937
审稿时长
29 weeks
期刊介绍:
Science Advances, an open-access journal by AAAS, publishes impactful research in diverse scientific areas. It aims for fair, fast, and expert peer review, providing freely accessible research to readers. Led by distinguished scientists, the journal supports AAAS's mission by extending Science magazine's capacity to identify and promote significant advances. Evolving digital publishing technologies play a crucial role in advancing AAAS's global mission for science communication and benefitting humankind.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


