静态三维结构决定了间期染色体远端位点对之间的快速动力学
IF 12.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
活细胞成像实验表明,增强子和启动子之间的远端动力学出乎意料地迅速,与标准聚合物模型不兼容。紧凑的静态染色质组织与动态染色质之间的不一致是一个难题,违反了预期的结构-功能关系。我们开发了一种理论,通过准确地从静态Hi-C接触图或固定细胞成像数据确定三维(3D)结构来预测染色质动力学。利用计算的三维坐标,该理论准确地预测了实验观察到的两点染色质动力学。它预测了快速的增强子-启动子相互作用,并揭示了两点松弛时间和基因组分离之间的比例关系,与最近的测量结果密切匹配。该理论预测,内聚蛋白耗竭加速了单位点扩散,同时显著减缓了拓扑相关域内的弛豫动力学。我们的研究结果表明,可以从静态结构数据中可靠地推断出染色质动力学,从而加强了三维染色质结构支配动态行为的概念。这个总体框架为探索不同生物背景下的染色质动力学提供了强大的工具。本文章由计算机程序翻译,如有差异,请以英文原文为准。
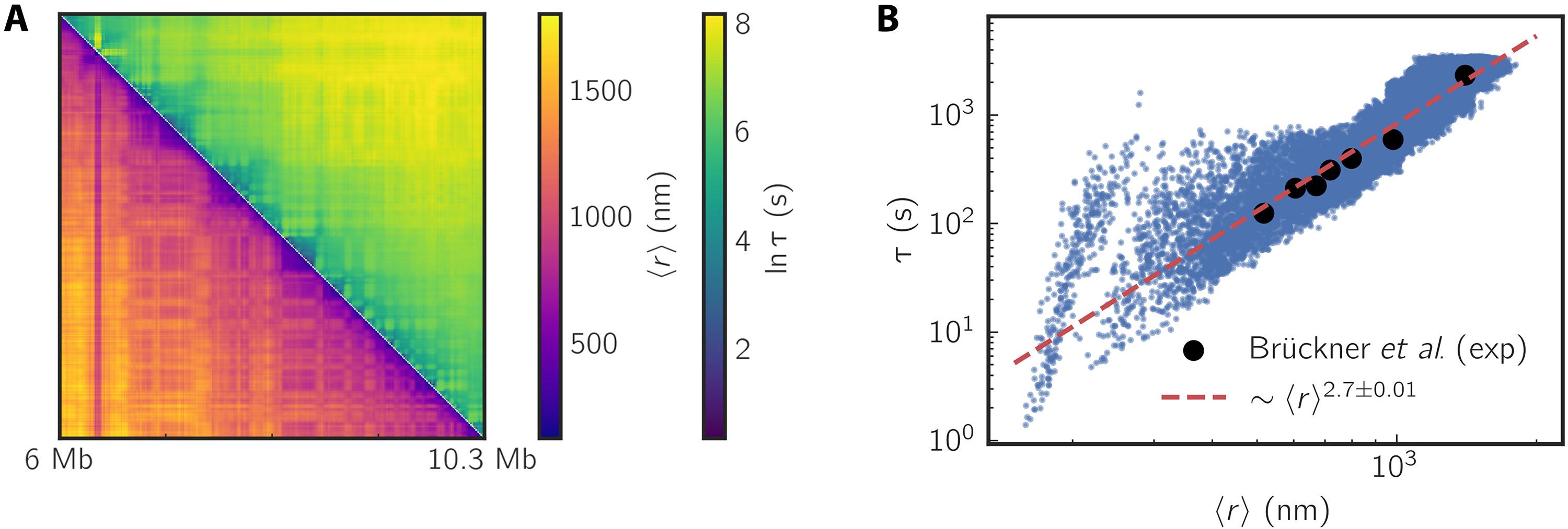
Static three-dimensional structures determine fast dynamics between distal loci pairs in interphase chromosomes
Live-cell imaging experiments have shown that the distal dynamics between enhancers and promoters are unexpectedly rapid and incompatible with standard polymer models. The discordance between the compact static chromatin organization and dynamics is a conundrum that violates the expected structure–function relationship. We developed a theory to predict chromatin dynamics by accurately determining three-dimensional (3D) structures from static Hi-C contact maps or fixed-cell imaging data. Using the calculated 3D coordinates, the theory accurately forecasts experimentally observed two-point chromatin dynamics. It predicts rapid enhancer–promoter interactions and uncovers a scaling relationship between two-point relaxation time and genomic separation, closely matching recent measurements. The theory predicts that cohesin depletion accelerates single-locus diffusion while significantly slowing relaxation dynamics within topologically associating domains. Our results demonstrate that chromatin dynamics can be reliably inferred from static structural data, reinforcing the notion that 3D chromatin structure governs dynamic behavior. This general framework offers powerful tools for exploring chromatin dynamics across diverse biological contexts.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Science Advances
综合性期刊-综合性期刊
CiteScore
21.40
自引率
1.50%
发文量
1937
审稿时长
29 weeks
期刊介绍:
Science Advances, an open-access journal by AAAS, publishes impactful research in diverse scientific areas. It aims for fair, fast, and expert peer review, providing freely accessible research to readers. Led by distinguished scientists, the journal supports AAAS's mission by extending Science magazine's capacity to identify and promote significant advances. Evolving digital publishing technologies play a crucial role in advancing AAAS's global mission for science communication and benefitting humankind.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


