伪tpase介导的蛋白-蛋白相互作用的结构基础
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
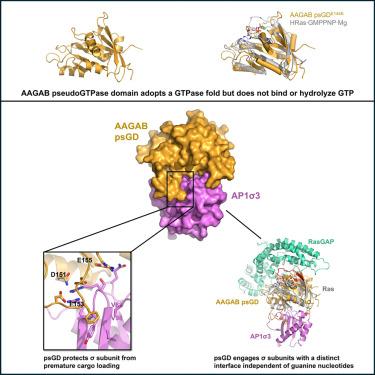
gtpase通过鸟嘌呤核苷酸结合引发的构象变化调节细胞过程。最近,从细菌到人类,已经发现了gtpase的催化无活性对应物——伪gtpase,尽管它们的功能和机制仍未被探索。在这里,我们利用生物化学和x射线晶体学证明了装配伴侣AAGAB的n端区域是一类伪tpase。此外,我们发现AAGAB伪伪tpase结构域(psGD)与AP1和AP2接头复合物的σ亚基相互作用,AP1和AP2接头复合物是参与网格蛋白介导的膜运输的异四聚体复合物。AAGAB psGD通过与传统GTPase相互作用区域不同的独特界面接合σ亚基。进一步的生化和基于细胞的分析证实了新发现的界面在结合和膜运输中的关键作用。总之,我们的研究结果确定AAGAB伪tpase结构域是一个关键的蛋白质-蛋白质相互作用模块。这些发现为研究伪tpase的结构基础和分子机制提供了新的思路。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural basis of pseudoGTPase-mediated protein-protein interactions
GTPases regulate cellular processes through conformational changes triggered by guanine nucleotide binding. Recently, pseudoGTPases, the catalytically inactive counterparts of GTPases, have been identified across species from bacteria to human, although their functions and mechanisms remain unexplored. Here, we demonstrate that the N-terminal region of the assembly chaperone AAGAB is a class I pseudoGTPase using biochemistry and X-ray crystallography. Furthermore, we discovered that the AAGAB pseudoGTPase domain (psGD) interacts with the σ subunits of AP1 and AP2 adaptor complexes, which are heterotetrameric complexes involved in clathrin-mediated membrane trafficking. AAGAB psGD engages the σ subunits via a unique interface distinct from the conventional GTPase interacting regions. Further biochemical and cell-based assays confirmed the crucial role of the newly identified interface in binding and membrane trafficking. Collectively, our results establish AAGAB pseudoGTPase domain as a critical protein-protein interaction module. These findings offer new insight into the structural basis and molecular mechanisms of pseudoGTPases.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


