紫外线诱变增强了人肠球菌LD3的细菌素产生:遗传和结构表征
IF 0.9
Q4 GENETICS & HEREDITY
引用次数: 0
摘要
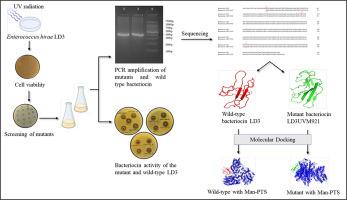
紫外辐射通常被用来诱导不同细菌基因组的突变,以了解代谢产物的结构-功能关系。我们之前已经从一种本土发酵食品中分离出产细菌素的hiraenterococcus LD3,并对其益生菌特性进行了表征。由于细菌素的产生是一种遗传现象,我们已经使用紫外线辐射产生突变体进行细菌素生产的表征。方法利用254 nm紫外辐射产生突变,并用细菌素的产生水平筛选突变体。利用MEGA-11和I-TASSER软件对细菌素基因进行扩增、测序、系统发育和结构分析。利用BIOVIA discovery studio可视化工具对野生型和突变型细菌素与目标甘露糖磷酸转移酶系统(Man-PTS)进行分子对接,了解其功能。结果突变株LD3UVM921的细菌素产量(320 AU/mL)高于野生株LD3 (160 AU/mL)。细菌素基因在紫外线照射后显示胞嘧啶残基缺失,鸟嘌呤为胸腺嘧啶,胞嘧啶为腺嘌呤。突变体LD3UVM921的二级结构为无α -螺旋、无β -链38.54%、无随机圈61.45%,而野生型为7.27%、无α -螺旋33.63%、无随机圈59.09%。分子对接分析表明,与野生型相比,突变型细菌素LD3UVM921与Man-PTS的结合亲和力更高,导致细菌素活性更高。结论紫外线照射成功地引起了细菌素基因的改变,并影响了其产生水平。该突变体可用于食品和制药工业中大规模生产细菌素。本文章由计算机程序翻译,如有差异,请以英文原文为准。
UV-induced mutagenesis enhances bacteriocin production in Enterococcus hirae LD3: Genetic and structural characterization
Background
Ultraviolet radiation is generally used to induce mutations in different bacterial genomes to understand structure-function relationship of metabolites. We have previously isolated bacteriocin-producing Enterococcus hirae LD3 from an indigenous fermented food and characterized for probiotic properties. Since bacteriocin production is a genetic phenomenon, we have used UV-radiation to generate mutants for bacteriocin production characterization.
Methods
Mutation was generated using UV radiation at 254 nm and mutants were screened using the level of bacteriocin production. Bacteriocin gene was amplified, sequenced, and phylogenetic and structure analysis was performed using MEGA-11 and I-TASSER software. The molecular docking of wild-type and mutant bacteriocins with target mannose phosphotransferase system (Man-PTS) was performed using BIOVIA discovery studio visualizer to understand the function.
Results
The mutant strain LD3UVM921 showed higher bacteriocin production (320 AU/mL) compared to wild-type strain LD3 (160 AU/mL). The bacteriocin gene showed a deletion of a cytosine residue and guanine to thymine and a cytosine to adenine transversion after UV exposure. The secondary structure of mutant bacteriocin LD3UVM921 was without alpha helix, 38.54 % beta-strand and 61.45 % random coil compared to 7.27 % alpha helix, 33.63 % beta-strand, and 59.09 % random coil in wild-type. The molecular docking analysis indicated higher binding affinity of mutant bacteriocin LD3UVM921 with Man-PTS compared to wild-type leading to higher bacteriocin activity.
Conclusion
The UV radiation successfully generated alteration in bacteriocin gene and affected its production level. The over-producing mutant of E. hirae LD3 may be used in food and pharmaceutical industries for large scale production of bacteriocin.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Gene Reports
Biochemistry, Genetics and Molecular Biology-Genetics
CiteScore
3.30
自引率
7.70%
发文量
246
审稿时长
49 days
期刊介绍:
Gene Reports publishes papers that focus on the regulation, expression, function and evolution of genes in all biological contexts, including all prokaryotic and eukaryotic organisms, as well as viruses. Gene Reports strives to be a very diverse journal and topics in all fields will be considered for publication. Although not limited to the following, some general topics include: DNA Organization, Replication & Evolution -Focus on genomic DNA (chromosomal organization, comparative genomics, DNA replication, DNA repair, mobile DNA, mitochondrial DNA, chloroplast DNA). Expression & Function - Focus on functional RNAs (microRNAs, tRNAs, rRNAs, mRNA splicing, alternative polyadenylation) Regulation - Focus on processes that mediate gene-read out (epigenetics, chromatin, histone code, transcription, translation, protein degradation). Cell Signaling - Focus on mechanisms that control information flow into the nucleus to control gene expression (kinase and phosphatase pathways controlled by extra-cellular ligands, Wnt, Notch, TGFbeta/BMPs, FGFs, IGFs etc.) Profiling of gene expression and genetic variation - Focus on high throughput approaches (e.g., DeepSeq, ChIP-Seq, Affymetrix microarrays, proteomics) that define gene regulatory circuitry, molecular pathways and protein/protein networks. Genetics - Focus on development in model organisms (e.g., mouse, frog, fruit fly, worm), human genetic variation, population genetics, as well as agricultural and veterinary genetics. Molecular Pathology & Regenerative Medicine - Focus on the deregulation of molecular processes in human diseases and mechanisms supporting regeneration of tissues through pluripotent or multipotent stem cells.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


