将蛋白质结合物扩散到内在无序的蛋白质上
IF 48.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
与内在无序蛋白(IDPs)和内在无序区(IDRs)结合的蛋白具有高亲和力和特异性,可用于治疗和诊断。然而,针对国内流离失所者或境内流离失所者的一般方法尚未制订。本研究表明,仅从输入的靶序列开始,并自由采样靶蛋白和结合蛋白构象,RFdiffusion5可以生成广泛构象的IDPs和idr的结合物。我们使用这种方法生成了不同构象的IDPs amylin, C-peptide, VP48和BRCA1_ARATH的结合物,其解离常数(Kd)范围为3至100 nM。对于IDRs G3BP1,常见的细胞因子受体γ-链(IL-2RG)和朊病毒蛋白,我们将结合物扩散到靶标的β-链构象上,获得了10到100 nM之间的Kd。荧光成像实验表明,这些结合剂与细胞中各自的靶标结合。G3BP1结合剂破坏细胞中应激颗粒的形成,而胰淀素结合剂抑制淀粉样纤维的形成并解离现有纤维,使溶酶体能够靶向单体和纤维状胰淀素,并提高基于质谱的胰淀素检测的灵敏度。我们的方法对于创建灵活的idp或idr的粘合剂应该是有用的,这些粘合剂跨越了广泛的内在构象偏好。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Diffusing protein binders to intrinsically disordered proteins
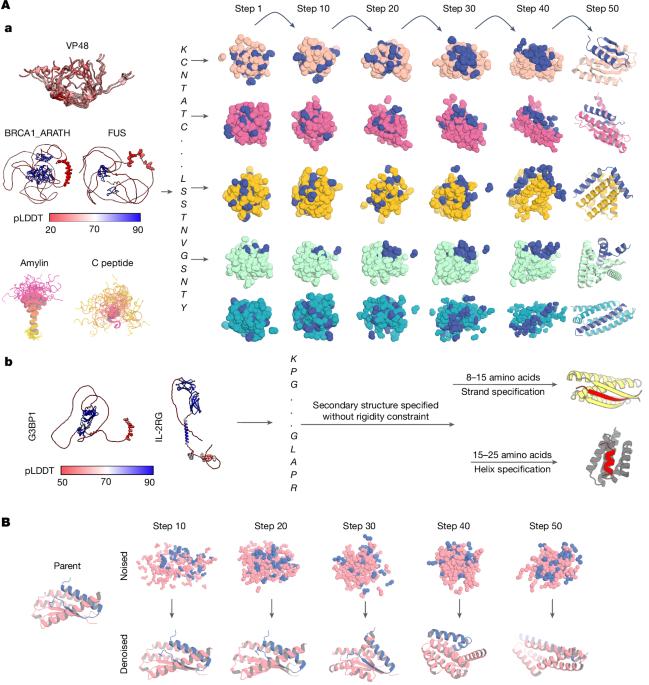
Proteins that bind to intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) with high affinity and specificity could be useful for therapeutic and diagnostic applications1–4. However, a general methodology for targeting IDPs or IDRs has yet to be developed. Here we show that starting only from the target sequence of the input, and freely sampling both target and binding protein conformations, RFdiffusion5 can generate binders to IDPs and IDRs in a wide range of conformations. We used this approach to generate binders to the IDPs amylin, C-peptide, VP48 and BRCA1_ARATH in diverse conformations with a dissociation constant (Kd) ranging from 3 to 100 nM. For the IDRs G3BP1, common cytokine receptor γ-chain (IL-2RG) and prion protein, we diffused binders to β-strand conformations of the targets, obtaining Kd between 10 and 100 nM. Fluorescence imaging experiments show that the binders bind to their respective targets in cells. The G3BP1 binder disrupts stress granule formation in cells, and the amylin binder inhibits amyloid fibril formation and dissociates existing fibres, enables targeting of both monomeric and fibrillar amylin to lysosomes, and increases the sensitivity of mass spectrometry-based amylin detection. Our approach should be useful for creating binders to flexible IDPs or IDRs spanning a wide range of intrinsic conformational preferences. Using RFdiffusion, a general method for targeting intrinsically disordered proteins and regions for protein design has been developed.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


