一个完整的单核和空间转录组图谱揭示了人类海马的分子景观
IF 20
1区 医学
Q1 NEUROSCIENCES
引用次数: 0
摘要
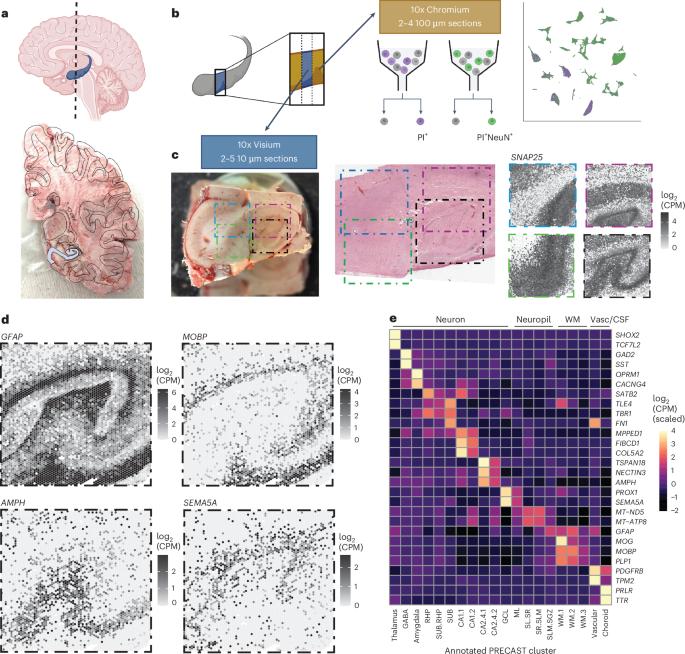
海马中具有独特形态、生理和连接的细胞类型具有与认知和情绪相关的特殊功能。这些细胞类型在空间上是有组织的,因此需要保留细胞结构组织的分子分析策略。在这里,我们从10个成年神经典型供体的人类海马前部生成了空间分辨转录组学(SRT)和单核rna测序(snRNA-seq)数据。利用非负矩阵分解(NMF)和标签转移,我们通过定义snRNA-seq数据中的基因表达模式,然后推断SRT数据中的表达来整合这些数据。这些模式捕获了神经元细胞类型的转录变异,并表明了兴奋性和抑制性突触后特化的空间组织。利用啮齿类动物数据集的NMF和标签转移方法,我们确定了人类SRT数据集中活动依赖性转录和电路连接的假设模式。最后,我们表征了与锥体神经元相对应的NMF模式的空间组织,并鉴定了海马后、枕下和枕前的区域特异性snRNA-seq簇。为了使这个分子图谱被广泛访问,原始和处理过的数据是免费提供的,包括通过交互式web应用程序。本文章由计算机程序翻译,如有差异,请以英文原文为准。

An integrated single-nucleus and spatial transcriptomics atlas reveals the molecular landscape of the human hippocampus
Cell types in the hippocampus with unique morphology, physiology and connectivity serve specialized functions associated with cognition and mood. These cell types are spatially organized, necessitating molecular profiling strategies that retain cytoarchitectural organization. Here we generated spatially-resolved transcriptomics (SRT) and single-nucleus RNA-sequencing (snRNA-seq) data from anterior human hippocampus in ten adult neurotypical donors. Using non-negative matrix factorization (NMF) and label transfer, we integrated these data by defining gene expression patterns within the snRNA-seq data and then inferring expression in the SRT data. These patterns captured transcriptional variation across neuronal cell types and indicated spatial organization of excitatory and inhibitory postsynaptic specializations. Leveraging the NMF and label transfer approach with rodent datasets, we identified putative patterns of activity-dependent transcription and circuit connectivity in the human SRT dataset. Finally, we characterized the spatial organization of NMF patterns corresponding to pyramidal neurons and identified regionally-specific snRNA-seq clusters of the retrohippocampus, subiculum and presubiculum. To make this molecular atlas widely accessible, raw and processed data are freely available, including through interactive web applications. The topographical organization of cells in the hippocampus reflects its ability to regulate mood and cognition. Here the authors generate a spatially resolved gene expression map in the human hippocampus to enable cross-species and functional interpretation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature neuroscience
医学-神经科学
CiteScore
38.60
自引率
1.20%
发文量
212
审稿时长
1 months
期刊介绍:
Nature Neuroscience, a multidisciplinary journal, publishes papers of the utmost quality and significance across all realms of neuroscience. The editors welcome contributions spanning molecular, cellular, systems, and cognitive neuroscience, along with psychophysics, computational modeling, and nervous system disorders. While no area is off-limits, studies offering fundamental insights into nervous system function receive priority.
The journal offers high visibility to both readers and authors, fostering interdisciplinary communication and accessibility to a broad audience. It maintains high standards of copy editing and production, rigorous peer review, rapid publication, and operates independently from academic societies and other vested interests.
In addition to primary research, Nature Neuroscience features news and views, reviews, editorials, commentaries, perspectives, book reviews, and correspondence, aiming to serve as the voice of the global neuroscience community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


