淀粉样多形体的结构和热力学分类
IF 4.3
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
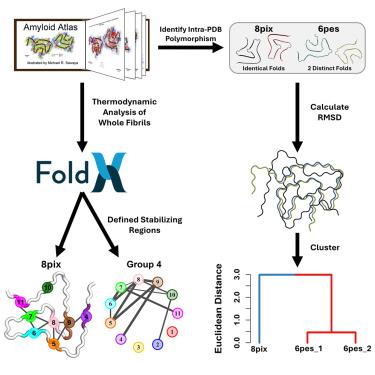
到目前为止,超过500个淀粉样蛋白结构已经达到接近原子的分辨率。这突出了纤维结构的巨大多样性,符合典型的交叉β淀粉样蛋白折叠。以α-突触核蛋白和tau淀粉样蛋白结构为模型,我们发现它们可以分层聚集成拓扑结构不同的折叠家族。尽管它们的拓扑结构不同,但原纤维显示出非常相似的能量分布,正如FoldX所确定的那样,相同的区域在不同的多态性中提供了稳定性。我们发现稳定淀粉样蛋白核心的区域以不同的方式产生不同的拓扑结构。结果提供了一个框架来分类新解决的纤维结构属于一个现有的类或形成一个新的拓扑交叉-β折叠。此外,该分析有助于比较疾病中发现的原纤维和体外形成的原纤维,包括它们最近的结构邻居。工作流程是自动化的,使用户能够在出现新的淀粉样蛋白结构时进行查询。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Structural and thermodynamic classification of amyloid polymorphs
Over 500 amyloid structures have been solved to date to near-atomic resolution. This has highlighted an enormous diversity of fibril structures conforming to the canonical cross-β amyloid fold. Using α-synuclein and tau amyloid structures as models, we show that they can be hierarchically clustered into topologically distinct fold families. Despite their different topologies, fibrils display remarkably similar energy profiles, as determined by FoldX, with the same regions providing stability among different polymorphs. We found that the regions that stabilize the amyloid core pair in different ways to generate distinct topologies. The results provide a framework to classify newly solved fibril structures as belonging to an existing class or forming a new topological cross-β fold. Furthermore, the analysis facilitates comparisons between fibrils found in disease and those formed in vitro, including their nearest structural neighbors. The workflow has been automated, enabling users to interrogate new amyloid structures as they emerge.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


