通过DNA测序大规模平行免疫肽丘提供了对癌症抗原呈递的见解
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
摘要
人类白细胞抗原(hla)是由人类基因组中最多的多态性基因编码的。HLA I类等位基因控制T细胞识别的抗原呈递,这是自身免疫、传染病和癌症的关键。目前关于HLA结合肽的知识是有限的,偏斜的,并且缺乏针对高价值靶点的人群范围的HLA结合谱。在这里,我们提出ESCAPE-seq(增强单链抗原呈递测序),这是一个大规模平行平台,通过深度DNA测序全面筛选I类hla -肽组合以供抗原呈递。ESCAPE-seq具有可编程性、高通量、敏感性和指定的病毒和癌症表位。我们同时评估了超过75,000种多肽- hla组合,揭示了广泛存在的致癌驱动突变表位,以及覆盖90%人口的不同HLA-A、HLA-B和HLA-C等位基因的融合。我们进一步鉴定了不同呈现的表位,比较了致癌热点突变与野生型。ESCAPE-seq能够一次性发现人群范围内的抗原呈递,为HLA特异性和基因组突变的免疫识别提供见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
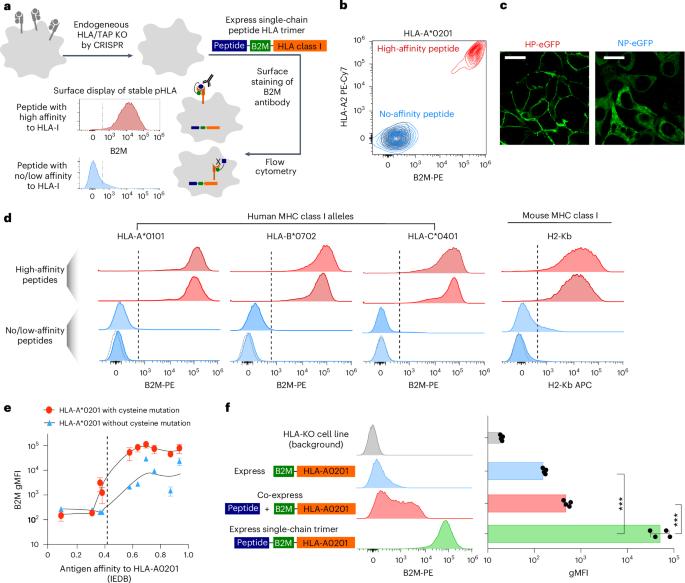
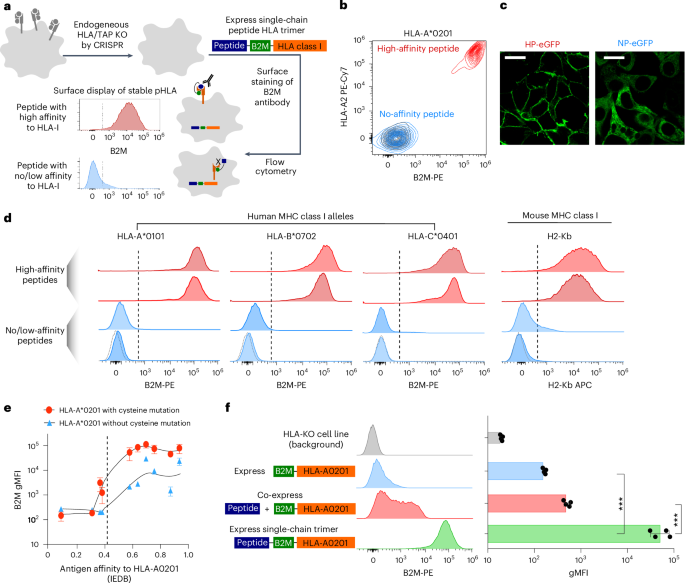
Massively parallel immunopeptidome by DNA sequencing provides insights into cancer antigen presentation
Human leukocyte antigens (HLAs) are encoded by the most polymorphic genes in the human genome. HLA class I alleles control antigen presentation for T cell recognition, which is pivotal for autoimmunity, infectious diseases and cancer. Current knowledge of HLA-bound peptides is limited, skewed and falls short of population-wide HLA binding profiles for high-value targets. Here we present ESCAPE-seq (enhanced single-chain antigen presentation sequencing), a massively parallel platform for comprehensive screening of class I HLA–peptide combinations for antigen presentation via deep DNA sequencing. ESCAPE-seq demonstrates programmability, high throughput, sensitivity and nominated viral and cancer epitopes. We simultaneously assessed over 75,000 peptide–HLA combinations, revealing broadly presented epitopes from oncogenic driver mutations and fusions across diverse HLA-A, HLA-B and HLA-C alleles that cover 90% of the human population. We further identified epitopes that are differentially presented, comparing oncogenic hotspot mutations versus wild type. ESCAPE-seq enables one-shot population-wide antigen presentation discovery, offering insights into HLA specificity and immune recognition of genomic mutations. ESCAPE-seq (enhanced single-chain antigen presentation sequencing) is a massively parallel platform for screening of class I HLA–peptide combinations for antigen presentation. The authors assess more than 75,000 peptide–HLA combinations, revealing presented epitopes from oncogenic driver mutations and fusions across diverse HLA-A, HLA-B and HLA-C alleles.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


