宿主内细菌进化和致病性的出现
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
利用全基因组测序监测细菌病原体,为了解其在宿主内的进化提供了重要见解,揭示了导致抗生素耐药性、免疫逃避表型和适应出现的致突变和选择性过程,从而实现了持续的人际传播。宿主内病原体种群的深度基因组和宏基因组测序也增强了我们追踪细菌传播的能力,这是感染控制的关键组成部分。这篇综述讨论了驱动人类细菌进化的主要过程,包括致病性和共生物种。最初,突变过程,包括突变特征如何揭示病原体生物学,以及选择压力驱动进化被考虑。水平基因转移和宿主内病原体竞争的动态也进行了研究,随后重点关注细菌发病机制的出现。最后,综述着重于宿主内遗传多样性在追踪细菌传播中的重要性及其对传染病控制和公共卫生的影响。本文章由计算机程序翻译,如有差异,请以英文原文为准。
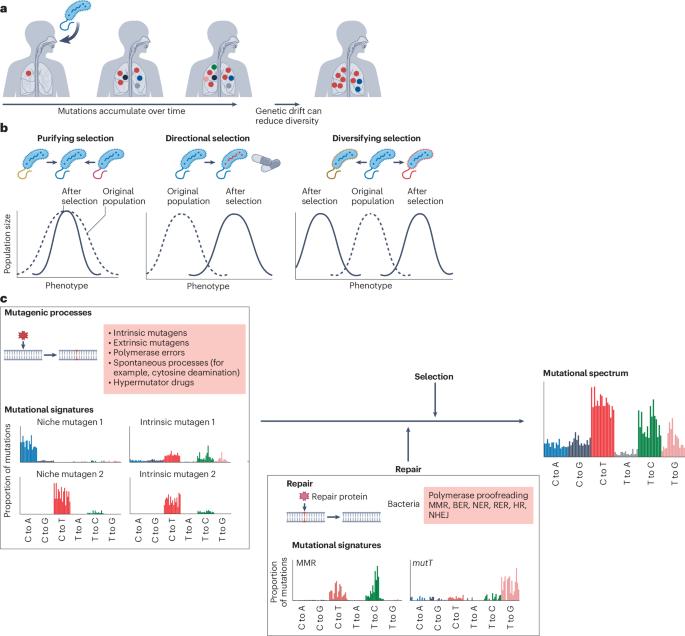

Within-host bacterial evolution and the emergence of pathogenicity
The use of whole-genome sequencing to monitor bacterial pathogens has provided crucial insights into their within-host evolution, revealing mutagenic and selective processes driving the emergence of antibiotic resistance, immune evasion phenotypes and adaptations that enable sustained human-to-human transmission. Deep genomic and metagenomic sequencing of intra-host pathogen populations is also enhancing our ability to track bacterial transmission, a key component of infection control. This Review discusses the major processes driving bacterial evolution within humans, including both pathogenic and commensal species. Initially, mutational processes, including how mutational signatures reveal pathogen biology, and the selective pressures driving evolution are considered. The dynamics of horizontal gene transfer and intra-host pathogen competition are also examined, followed by a focus on the emergence of bacterial pathogenesis. Finally, the Review focuses on the importance of within-host genetic diversity in tracking bacterial transmission and its implications for infectious disease control and public health. In this Review, Tonkin-Hill et al. discuss the processes driving bacterial evolution and emergence of pathogenesis within hosts, the importance of understanding within-host genetic diversity, and the implications for transmission analysis and infectious disease control.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


