基因组解析的长读测序扩展了已知的陆地栖息地微生物多样性
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
高通量、长读DNA测序的出现使微生物基因组从环境样本中大规模恢复成为可能。然而,由于这些环境的巨大复杂性,扩大陆地微生物基因组目录一直具有挑战性。在这里,我们对在Microflora Danica项目中收集的154个土壤和沉积物样本进行了深度、长读纳米孔测序,得到了15,314个以前未描述的微生物物种的基因组,使用我们定制的mmlong2工作流程进行了恢复。恢复的微生物基因组跨越1,086个以前未表征的属,并将原核生物生命树的系统发育多样性扩大了8%。长读组装还使数千个完整的核糖体RNA操纵子、生物合成基因簇和CRISPR-Cas系统得以恢复。此外,将恢复的基因组纳入公共基因组数据库大大提高了土壤和沉积物宏基因组数据集的物种水平分类率。这些发现表明,长读测序可以从高度复杂的生态系统中经济高效地恢复高质量的微生物基因组,这些生态系统仍然是生物多样性的未开发来源。本文章由计算机程序翻译,如有差异,请以英文原文为准。

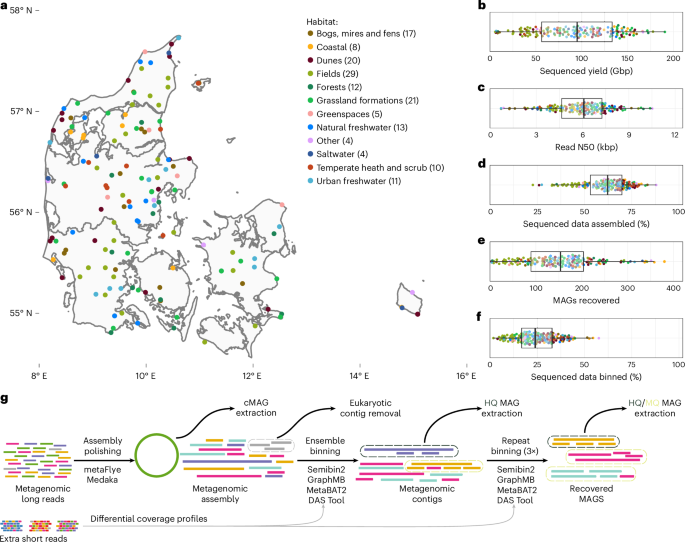
Genome-resolved long-read sequencing expands known microbial diversity across terrestrial habitats
The emergence of high-throughput, long-read DNA sequencing has enabled recovery of microbial genomes from environmental samples at scale. However, expanding the terrestrial microbial genome catalogue has been challenging due to the enormous complexity of these environments. Here we performed deep, long-read Nanopore sequencing of 154 soil and sediment samples collected during the Microflora Danica project, yielding genomes of 15,314 previously undescribed microbial species, recovered using our custom mmlong2 workflow. The recovered microbial genomes span 1,086 previously uncharacterized genera and expand the phylogenetic diversity of the prokaryotic tree of life by 8%. The long-read assemblies also enabled the recovery of thousands of complete ribosomal RNA operons, biosynthetic gene clusters and CRISPR-Cas systems. Furthermore, the incorporation of the recovered genomes into public genomic databases substantially improved species-level classification rates for soil and sediment metagenomic datasets. These findings demonstrate that long-read sequencing allows cost-effective recovery of high-quality microbial genomes from highly complex ecosystems, which remain an untapped source of biodiversity. Nanopore sequencing of Danish soils and sediments yields genomes from over 15,000 microbial species, expanding the phylogenetic diversity of prokaryotes by 8%.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


