基于单细胞图像的屏幕识别埃博拉病毒感染动态的宿主调节因子
IF 19.4
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
摘要
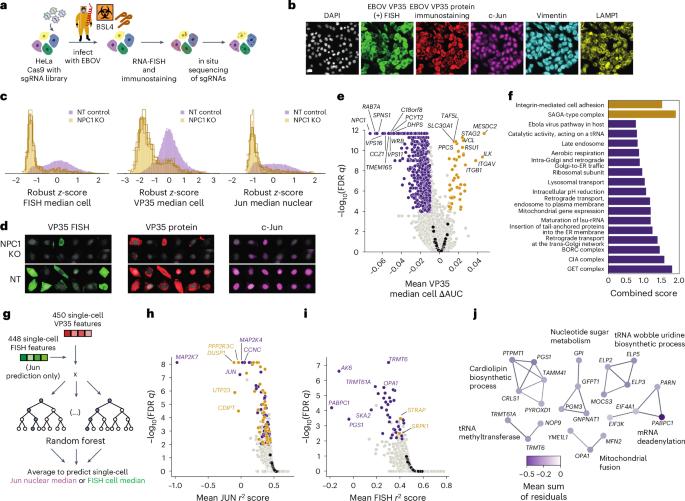
埃博拉病毒(EBOV)等丝状病毒引起频繁流行,病死率高,而治疗选择仍然有限。早期旨在确定EBOV潜在药物靶点的基因筛选依赖于可能无法完全概括病毒生命周期的系统。在这里,我们应用基于图像的全基因组CRISPR筛选,在39,085,093个细胞中鉴定出EBOV感染的998个宿主调节因子。深度学习模型将每个宿主因子与不同的病毒复制步骤联系起来。由此,我们证实了UQCRB是EBOV RNA复制的进入后调节剂,并表明小分子UQCRB抑制降低了体外病毒感染。使用随机森林模型,我们发现对剪接体相关因子(STRAP)的扰动破坏了病毒RNA和蛋白质之间的平衡。STRAP与病毒RNA加工蛋白VP35相关。这种全基因组筛选加上12个二级筛选,包括苏丹病毒和马尔堡病毒的验证实验,为病毒复制的宿主调节因子和治疗干预的潜在靶点提供了丰富的资源。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Single-cell image-based screens identify host regulators of Ebola virus infection dynamics
Filoviruses such as Ebola virus (EBOV) give rise to frequent epidemics with high case fatality rates while therapeutic options remain limited. Earlier genetic screens aimed to identify potential drug targets for EBOV relied on systems that may not fully recapitulate the virus life cycle. Here we applied an image-based genome-wide CRISPR screen to identify 998 host regulators of EBOV infection in 39,085,093 cells. A deep learning model associated each host factor with a distinct viral replication step. From this we confirmed UQCRB as a post-entry regulator of EBOV RNA replication and show that small-molecule UQCRB inhibition reduced virus infection in vitro. Using a random forest model, we found that perturbations on STRAP (a spliceosome-associated factor) disrupted the equilibrium between viral RNA and protein. STRAP was associated with VP35, a viral RNA processing protein. This genome-wide screen coupled with 12 secondary screens including validation experiments with Sudan and Marburg virus, presents a rich resource for host regulators of virus replication and potential targets for therapeutic intervention. A single-cell resolution image-based genome-wide CRISPR screen, analysed with deep learning and random forest models, identified host factors regulating Ebola virus infection at distinct stages in the viral life cycle.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


